Back to Journals » International Journal of General Medicine » Volume 14
HOTAIR Facilitates Endocrine Resistance in Breast Cancer Through ESR1/miR-130b-3p Axis: Comprehensive Analysis of mRNA-miRNA-lncRNA Network
Authors Zhang M, Wu K, Zhang P, Qiu Y, Bai F, Chen H
Received 20 May 2021
Accepted for publication 27 July 2021
Published 18 August 2021 Volume 2021:14 Pages 4653—4663
DOI https://doi.org/10.2147/IJGM.S320998
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Mingdi Zhang, Kejin Wu, Peng Zhang, Yiran Qiu, Fang Bai, Hongliang Chen
Department of Breast Surgery, Obstetrics and Gynecology Hospital of Fudan University, Shanghai, People’s Republic of China
Correspondence: Hongliang Chen
Department of Breast Surgery, Obstetrics and Gynecology Hospital of Fudan University, 419 Fangxie Road, Shanghai, 200011, People’s Republic of China
Tel/Fax +86-21-33189900
Email [email protected]
Background: To summarize the regulatory role of mRNA-miRNA-lncRNA network associated with endocrine therapy resistance (ETR) in breast cancer.
Methods: We analyzed the differentially expressed genes (DEGs), differentially expressed lncRNAs (DELs), and differentially expressed miRNAs (DEMs) in long-term estrogen-deprived (LTED) estrogen receptor (ER)-positive breast cancer cells (LTED MCF7) (modeling relapse on endocrine therapy) and MCF7 cells in the presence of estrogen (E2) (modeling a patient at primary diagnosis) by mining GSE120929 and GSE120930 datasets. The mRNA-miRNA-lncRNA network was constructed by multiple bioinformatic tools. The prognosis of genes from the network was validated in breast cancer patients with following systemic treatment (endocrine therapy) by GEPIA, Kaplan–Meier plotter and UALCAN database.
Results: Totally, 769 DEGs, 33 DEMs, and 10 DELs were selected. The mRNA-miRNA-lncRNA network was established including 60 mRNA nodes, 6 miRNA nodes and 3 lncRNA nodes. A significant module containing 3 nodes and 3 edges was calculated based on the mRNA-miRNA-lncRNA network. The hub genes in the network are ABCG2, ESR1 and GJA1. ESR1/miR-130b-3p/HOTAIR are significantly correlated with the prognosis of breast cancer patients with endocrine therapy.
Conclusion: This study provides a novel ETR-related mRNA-miRNA-lncRNA network. Further, we suggest that ESR1/miR-130b-3p/HOTAIR may be promising targets for clinical treatment of endocrine therapy-resistant breast cancer.
Keywords: endocrine therapy resistance, breast cancer, bioinformatic analysis, mRNA-miRNA-lncRNA network, GEO
Introduction
Breast cancer is the most common malignancy and the main cause of cancer death among women worldwide.1 Seventy-five percent of breast cancer tissues express estrogen receptor-alpha (ERα). Endocrine therapy (ET) is one of the most effective adjuvant therapies for these ER-positive patients,2–4 including selective estrogen receptor modulators (SERMs), selective estrogen receptor degraders (SERDs) and inhibitors of the enzyme aromatase (AI) converting androgens to estrogens.4,5 Clinically, more than half of the patients benefit from ET initially, however nearly 40% experience de novo or acquired ET resistance (ETR).6 Therefore, it is imperative to explore the potential mechanism of endocrine-resistant breast cancer and to identify novel therapeutic targets.
Previous studies have supported that noncoding RNA including miRNA, long noncoding RNA (lncRNA) and circular RNA (circRNA) are involved in the occurrence and development of multiple tumors.7,8 A competing endogenous RNA (ceRNA) hypothesis point out that mRNA, miRNA and lncRNA could achieve cross-talk between each other through a regulatory network.9 By the use of miRNA response elements, lncRNAs act as ‘sponges’ for miRNAs and result in the miRNAs-regulated mRNA levels alteration. The aberrant regulation of mRNA-miRNA-lncRNA network play key role in a variety of biological processes and molecular mechanism of tumors, including regulating gene transcription and post-transcriptional translation, epithelial-to-mesenchymal transition, signaling pathways, prognostic evaluation and therapeutic targets.10–13 The deregulation of protein‐coding and noncoding genes in endocrine resistance in luminal breast cancer has reported in several studies.14–16 However, understanding of the core mRNA-miRNA-lncRNA networks associated with endocrine-resistant breast cancer is still limited.
In this study, bioinformatic analysis were conducted to explore differentially expressed genes (DEGs), differentially expressed miRNAs (DEMs), and differentially expressed lncRNAs (DELs) in long-term estrogen-deprived (LTED) ER-positive breast cancer cells, to establish a mRNA-miRNA-lncRNA regulatory network associated with endocrine resistance, and to mine potential therapy targets for overcoming endocrine resistance.
Materials and Methods
Data Resources and Differentially Expression Analysis
To identify DEGs, DEMs, and DELs in endocrine resistant breast cancer cells, gene expression dataset (GSE120929) and miRNA expression dataset (GSE120930) were obtained by searching the keywords of “breast cancer” and “endocrine resistance” in GEO database (http://www.ncbi.nlm.nih.gov/geo). |log2 fold change (FC)|>2 and p<0.05 was the cutoff criteria. Volcano plots were created to visualize the expression of all genes in the datasets.
Function Enrichment Analysis
Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed using Database for annotation, visualization, and integrated discovery (DAVID, https://david.ncifcrf.gov/). p < 0.05 and enrichment score> 1.0 were significant.
Comprehensive Analysis of PPI Network, Identification and Validation of Hub Genes
The protein–protein interaction (PPI) network was constructed using The Search Tool for the Retrieval of Interacting Genes (STRING) database and visualized by Cytoscape 3.6.1. The key modules in PPI network was selected using the cutoff criteria (MCODE score > 3) with the default parameters (degree cutoff = 2, node score cutoff = 0.2, K‐core = 2, and Max depth = 100). Cytoscape CentiScape was used to screen hub genes in the network, according to the degree of centrality. The expression levels of hub genes were further validated between breast cancer and normal samples in GEPIA database. The prognostic values of hub genes including overall survival (OS) and relapse-free survival (RFS) was evaluated in breast cancer patients and breast cancer patients with following systemic treatment (endocrine therapy) in GEPIA and Kaplan–Meier plotter database.
Identification of miRNA Target Genes and lncRNAs
The target genes interacted with miRNAs were predicted based on the miRTarbase database (http://miRTarBase.cuhk.edu.cn/). The upstream lncRNAs of miRNAs were predicted by miRNet database. “Organism-H.sapies”, “Breast cancerous tissues” and “target type-lncRNAs” were considered as selection criteria. FunRich 3.1 software was used to summarize the overlapping genes between the predicted miRNA targets and DEGs, the predicted lncRNAs and DELs.
Construction and Validation of mRNA-miRNA-lncRNA Regulatory Network
The interaction among mRNA, miRNA and lncRNA related to endocrine resistance was constructed according to the lncRNA targets and miRNA targets prediction using the Cytoscape software. The expression levels of miRNAs and hub genes from the network were further validated between breast cancer and normal samples in GEPIA and UALCAN database. The prognostic values of hub genes including OS and RFS was evaluated in breast cancer patients, ER-positive breast cancer patients and breast cancer patients with following systemic treatment (endocrine therapy) in GEPIA and Kaplan–Meier plotter database.
Results
Identification of DEGs, DEMs, and DELs
Dataset GSE120929 and GSE120930 were downloaded to select DEGs, DEMs and DELs in LTED ER-positive breast cancer cells (LTED MCF7) (modeling relapse on endocrine therapy) and MCF7 cells in the presence of estrogen (E2) (modeling a patient at primary diagnosis) (Figure 1A). A total of 769 DEGs including 443 up-regulated and 326 down-regulated genes were identified between LTED cells and MCF-7 cells with E2 (Figure 1B). A total of 33 DEMs, 18 upregulated and 15 downregulated miRNAs, were determined in GSE120930 (Figure 1C). As for DELs, 10 DELs (7 up-regulated and 3 down-regulated lncRNAs) were found in total (Figure 1D).
Functional Analysis for the Significant DEGs
For upregulated significant DEGs, the enriched GO functions are cell adhesion, cell–cell signaling and skeletal system development in the biological process (BP) category, calcium ion binding, calmodulin binding and heme binding in the molecular function (MF) category, and integral component of membrane, plasma membrane and integral component of plasma membrane in the cellular component (CC) category (Figure S1A–C). Besides, Figure S2A indicated that the upregulated significant DEGs were enriched in calcium signaling pathway, protein digestion and absorption and gap junction.
The downregulated significant DEGs were enriched in type I interferon signaling pathway, defense response to virus and response to virus in the BP category, cytokine activity, signal transducer activity and double-stranded RNA binding in the MF category, and plasma membrane, cytosol and extracellular space in the CC category (Figure S1D–F). Additionally, KEGG pathway enrichment analysis indicated that the downregulated DEGs were particularly involved in pentose and glucuronate interconversions, hepatitis C and Jak-STAT signaling pathway (Figure S2B).
Identification and Validation of Hub Genes
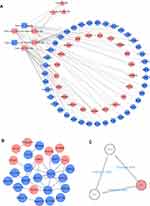
313 nodes and 768 edges of upregulated significant DEGs and 257 nodes and 290 edges of downregulated significant DEGs were mapped in the PPI network. The interaction among the top 30 upregulated and downregulated hub genes were visualized by Cytoscape software (Figure S3A–B). Two significant modules in the PPI network were identified using MCODE (Figure S3C and D). The top 10 hub genes and the corresponding node degrees are shown in Table 1 and the top 10 upregulated hub genes were CXCR4, ADCY1, CD44, ESR1, GNAI1, COL1A1, GCGR, CCR1, SOCS3 and PTGER3, and the top 10 downregulated hub genes were STAT1, IRF7, HERC6, IRF9, USP18, ISG15, IFIT1, HERC5, OAS1 and IFIT3.
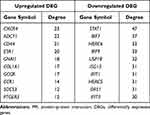 |
Table 1 The Top 10 Hub Genes in PPI Networks |
The expression levels and prognostic values of those key mRNAs in breast cancer were validated in the GEPIA database. Only 1 hub genes (ESR1) was significantly overexpressed and associated with worse OS of patients with breast cancer (Figure 2A and B). The prognostic values of those hub genes were also evaluated using Kaplan–Meier plotter database. As a result, only one of these hub genes (ESR1) showed both worse OS and RFS in breast cancer patients with following systemic treatment (endocrine therapy) (Figure 2C and D).
Integration of mRNA-miRNA-lncRNA Regulatory Network
To explore the role of miRNAs and lncRNAs in endocrine resistant cells, the mRNA-miRNA-lncRNA interaction network was constructed based on 60 mRNA nodes, 6 miRNA nodes and 3 lncRNA nodes (Figure 3A). Six overlapping DEMs (hsa-miR-130b-3p, hsa-miR-196a-5p, hsa-miR-23b-3p, hsa-miR-10a-5p, hsa-miR-195-5p and hsa-miR-370-3p) were identified in dataset GSE120930. The target genes of hsa-miR-130b-3p, hsa-miR-196a-5p, hsa-miR-23b-3p, hsa-miR-10a-5p, hsa-miR-195-5p and hsa-miR-370-3p overlapping with DEGs are listed in Table 2. The connections between DELs and lncRNAs predicted by these six miRNAs in miRNet database are shown in Table 3. The interaction of the 60 overlapping mRNA nodes were presented in the PPI network (Figure 3B). A significant module containing 3 nodes and 3 edges was identified. The hub genes were ABCG2, ESR1 and GJA1 (Figure 3C). In the network, we found ESR1, the target genes of hsa-miR-130b-3p belonging to the top 10 hub genes, revealing the potential role of ESR1/miR-130b-3p/HOTAIR in regulating endocrine resistance in breast cancer (Figure 3B and C).
 |
Table 2 Overlapping Genes Between the miRNA Targets and Overlapping DEGs |
 |
Table 3 Overlapping lncRNAs Between the miRNA Upstream Targets and Overlapping DELs |
GO analysis revealed that the 60 DEGs in network were enriched in 46 GO terms. The top enriched GO terms are response to hypoxia in BP terms, protein binding in MF terms, integral component of membrane in CC terms (Figure S4A–C). The top KEGG pathways were Pathways in cancer, Proteoglycans in cancer, HIF-1 signaling pathway (Figure S4D).
The Validation of Genes in mRNA-miRNA-lncRNA Network
The expression of miRNAs in the network and the prognostic values were assessed in breast cancer patients and breast cancer patients with following systemic treatment (endocrine therapy) in the UALCAN database and Kaplan–Meier plotter database, respectively. Three of these overlapping miRNAs (hsa-miR-130b-3p, hsa-miR-10a-5p and hsa-miR-195-5p) were aberrantly expressed in breast cancer and the expression was associated with tworse OS of breast cancer patients with following systemic treatment (endocrine therapy) (Figure 4). As for lncRNAs, HOTAIR is the only one significantly expressed genes from the network in breast cancer (Figure 5A). The high expression of HOTAIR was significantly related to worse OS in breast cancer patients and worse RFS in breast cancer patients with following systemic treatment (endocrine therapy), lightly related to worse OS in ER-positive patients (Figure 5B–D). Based on the hub genes selected from 60 overlapping mRNA nodes in the network (ABCG2, ESR1 and GJA1), ESR1 and ABCG2 showed aberrant expression related to OS in breast cancer patients (Figures 2 and S5A and B). High expression of ESR1 and low expression of GJA1 showed both worse OS and RFS in breast cancer patients with following systemic treatment (endocrine therapy) (Figures 2 and S5C and D).
 |
Figure 4 The expression and prognostic values of miR-130b (A and B), miR-10a (C and D) and miR-195 (E and F) from mRNA-miRNA-lncRNA regulatory network in UALCAN and Kaplan-Meier plotter databases. |
Discussion
ET remains the mainstream adjuvant treatment on hormone receptor-positive breast cancer, however, de novo or acquired ETR is a major limitation of treatment. It is essential to explore effective biomarkers predicting ETR and find alternative treatment on endocrine resistant tumors.
In our study, 769 DEGs, 33 DEMs, and 10 DELs were identified in the endocrine‐resistant cell by mining the datasets from the GEO database in order to set up ETR related mRNA-miRNA-lncRNA network including 60 mRNA s, 6 miRNAs and 3 lncRNAs. The role of noncoding RNAs in breast cancer and endocrine resistant breast cancers have been recently reviewed.15,17,18 Overexpression of lncRNA breast cancer anti-estrogen resistance 4 (BCAR4) in tamoxifen sensitive cells partly arrogates the anti-proliferative effects of tamoxifen.19 BCAR4 is also considered as a biomarker for increased cancer invasiveness and tamoxifen resistance.20 Long non-coding RNA UCA1 promoted tamoxifen resistance in breast cancer cells by regulating HIF1α via sponging miR-18a.14 lncRNA HOTAIR upregulation promotes ligand independent ER activities and contributes to tamoxifen resistance in breast cancer.15
Additionally, KEGG pathway enrichment analysis indicated that the enriched pathways primarily involved pathways in cancer, proteoglycans in cancer, HIF-1 signaling pathway. Besides, GO analysis and pathway analysis revealed that the significant DEGs were mostly enriched in response to hypoxia. It has been documented that hypoxia-inducible factors, HIF-1 play key roles in doxorubicin resistance of breast cancer.16
Subsequently, the target of hsa-miR-130b-3p were found in the top 10 hub genes in 769 DEGs, revealing the vital role of ESR1/miR-130b-3p/HOTAIR in regulating endocrine resistance. The prognostic values of the genes from the mRNA-miRNA-lncRNA network were validated in multiple databases. Eventually, ESR1/miR-130b-3p/HOTAIR were not only differentially expressed in patients with breast cancer but also significantly correlated with the prognosis of breast cancer patients with following systemic treatment (endocrine therapy).
miR-130b acts as an onco-miRNAs and promotes the progression of cancer.21,22 Meanwhile, the overexpression of miR-130b is a risk factor for poor prognosis in prostate cancer patients.23 On the contrary, few studies also identified that downregulation of miR-130b plays a role in stimulating progression and angiogenesis of prostate cancer.24 In addition, miR-130b is a target miRNA of CASC15 and CASC15/miR-130b axis might be a novel therapy for bladder cancer and non-small cell lung cancer patients.25 It was reported that miR-130b was downregulated in multidrug resistant ovarian cancer cells and restoration of miR-130b expression could sensitize cells to anticancer drugs.26
HOX transcript antisense RNA (HOTAIR) has been identified as the first lncRNA correlated to poor prognosis of breast cancer, taking part in EMT and maintenance of breast cancer stem cells (bCSCs).27,28 Based on the Estrogen Responsive Elements (EREs) elements in HOTAIR promoter, estradiol regulate HOTAIR expression in ER-positive breast cancer cells.29 HOTAIR is reported to be significantly upregulated in tamoxifen-resistant breast cancer cells, contributing to tamoxifen resistance by interacting with ER.30
Among the overlapping DEGs, ABCG2 and GJA1 were also screened as hub genes. The expression of ABCG2 was reported to be associated with triple-negative tumors.31 The ABCG2 signaling was involved in autophagy-promoting drug resistance in breast cancer stem cells.32 A recent study reported GJA1 is dysregulated in breast cancer and luminal tumors with high levels of GJA1 mRNA were associated with a better prognostic.33,34
According to the above analysis results, mRNA-miRNA-lncRNA regulatory network was built up. This is a novel mRNA-miRNA-lncRNA regulatory network reported in endocrine resistance. This study provides some powerful evidence for molecular mechanism of endocrine resistance. However, there are a few limitations in our study: the whole set of DEGs was extracted from cell lines and not from tumor tissues; the regulatory network is related to endocrine therapy resistance without specifying which category or which specific medicine. Further study will validate these findings by experiment on cell lines and human tissues.
Data Sharing Statement
The GEO data (GSE120929 and GSE120930) associated with the paper is available in Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/).
Consent for Publication
The details of any images, videos, recordings, etc can be published.
Funding
This work was supported by grants from clinical project of Shanghai Municipal Commission of Health (grant no. 20194Y0123 to MD Zhang), clinical cultivation project of Obstetrics and Gynecology Hospital affiliated to Fudan University (grant no. 20017 to MD Zhang), medical innovation research project of Science and Technology Innovation Action Plan (grant no. 20Y11914500 to MD Zhang).
Disclosure
The authors report no conflicts of interest in this work.
References
1. DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69(6):438–451. doi:10.3322/caac.21583
2. Colditz GA. Relationship between estrogen levels, use of hormone replacement therapy, and breast cancer. J Natl Cancer Inst. 1998;90(11):814–823. doi:10.1093/jnci/90.11.814
3. McDonnell DP, Norris JD. Connections and regulation of the human estrogen receptor. Science. 2002;296(5573):1642–1644. doi:10.1126/science.1071884
4. Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9(9):631–643. doi:10.1038/nrc2713
5. Najim O, Seghers S, Sergoynne L, et al. The association between type of endocrine therapy and development of estrogen receptor-1 mutation(s) in patients with hormone-sensitive advanced breast cancer: a systematic review and meta-analysis of randomized and non-randomized trials. Biochim Biophys Acta Rev Cancer. 2019;1872(2):188315. doi:10.1016/j.bbcan.2019.188315
6. Haque MM, Desai KV. Pathways to endocrine therapy resistance in breast cancer. Front Endocrinol (Lausanne). 2019;10:573. doi:10.3389/fendo.2019.00573
7. Fu XD. Non-coding RNA: a new frontier in regulatory biology. Natl Sci Rev. 2014;1(2):190–204. doi:10.1093/nsr/nwu008
8. Gao L, Li X, Nie X, et al. Construction of novel mRNA-miRNA-lncRNA regulatory networks associated with prognosis of ovarian cancer. J Cancer. 2020;11(23):7057–7072. doi:10.7150/jca.49557
9. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
10. Lu C, Luo X, Xing C, et al. Construction of a novel mRNA-miRNA-lncRNA network and identification of potential regulatory axis associated with prognosis in colorectal cancer liver metastases. Aging. 2021;13(11):14968–14988. doi:10.18632/aging.203049
11. Hu B, Ma X, Fu P, et al. miRNA-mRNA regulatory network and factors associated with prediction of prognosis in hepatocellular carcinoma. Genomics Proteomics Bioinformatics. 2021:S1672-0229(21)00059-0. doi:10.1016/j.gpb.2021.03.001
12. Landeros N, Santoro PM, Carrasco-Avino G, Corvalan AH. Competing endogenous RNA networks in the epithelial to mesenchymal transition in diffuse-type of gastric cancer. Cancers. 2020;12(10):10. doi:10.3390/cancers12102741
13. Chen X, Xu J, Zeng F, et al. Inferring cell subtypes and LncRNA function by a cell-specific ceRNA network in breast cancer. Front Oncol. 2021;11:656675. doi:10.3389/fonc.2021.656675
14. Garcia-Becerra R, Santos N, Diaz L, Camacho J. Mechanisms of resistance to endocrine therapy in breast cancer: focus on signaling pathways, miRNAs and genetically based resistance. Int J Mol Sci. 2012;14(1):108–145. doi:10.3390/ijms14010108
15. Muluhngwi P, Klinge CM. Roles for miRNAs in endocrine resistance in breast cancer. Endocr Relat Cancer. 2015;22(5):R279–R300. doi:10.1530/ERC-15-0355
16. Zhou J, Teng R, Wang Q, et al. Endocrine resistance in breast cancer: current status and a perspective on the roles of miRNAs. Oncol Lett. 2013;6(2):295–305. doi:10.3892/ol.2013.1405
17. Hayes EL, Lewis-Wambi JS. Mechanisms of endocrine resistance in breast cancer: an overview of the proposed roles of noncoding RNA. Breast Cancer Res. 2015;17(1):40. doi:10.1186/s13058-015-0542-y
18. van Schooneveld E, Wildiers H, Vergote I, Vermeulen PB, Dirix LY, Van Laere SJ. Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Res. 2015;17(1):21. doi:10.1186/s13058-015-0526-y
19. Godinho MF, Sieuwerts AM, Look MP, et al. Relevance of BCAR4 in tamoxifen resistance and tumour aggressiveness of human breast cancer. Br J Cancer. 2010;103(8):1284–1291. doi:10.1038/sj.bjc.6605884
20. Meijer D, van Agthoven T, Bosma PT, Nooter K, Dorssers LC. Functional screen for genes responsible for tamoxifen resistance in human breast cancer cells. Mol Cancer Res. 2006;4(6):379–386. doi:10.1158/1541-7786.MCR-05-0156
21. Chen H, Yang Y, Wang J, Shen D, Zhao J, Yu Q. miR-130b-5p promotes proliferation, migration and invasion of gastric cancer cells via targeting RASAL1. Oncol Lett. 2018;15(5):6361–6367.
22. Liu X, Kong C, Zhang Z. miR-130b promotes bladder cancer cell proliferation, migration and invasion by targeting VGLL4. Oncol Rep. 2018;39(5):2324–2332.
23. Hashimoto Y, Shiina M, Dasgupta P, et al. Upregulation of miR-130b contributes to risk of poor prognosis and racial disparity in African-American prostate cancer. Cancer Prev Res. 2019;12(9):585–598. doi:10.1158/1940-6207.CAPR-18-0509
24. Mu HQ, He YH, Wang SB, et al. MiR-130b/TNF-alpha/NF-kappaB/VEGFA loop inhibits prostate cancer angiogenesis. Clin Transl Oncol. 2020;22(1):111–121. doi:10.1007/s12094-019-02217-5
25. Yu DJ, Zhong M, Wang WL. Long noncoding RNA CASC15 is upregulated in non-small cell lung cancer and facilitates cell proliferation and metastasis via targeting miR-130b-3p. Eur Rev Med Pharmacol Sci. 2019;23(18):7943–7949.
26. Yang C, Cai J, Wang Q, et al. Epigenetic silencing of miR-130b in ovarian cancer promotes the development of multidrug resistance by targeting colony-stimulating factor 1. Gynecol Oncol. 2012;124(2):325–334. doi:10.1016/j.ygyno.2011.10.013
27. Mozdarani H, Ezzatizadeh V, Rahbar Parvaneh R. The emerging role of the long non-coding RNA HOTAIR in breast cancer development and treatment. J Transl Med. 2020;18(1):152. doi:10.1186/s12967-020-02320-0
28. Xu S, Kong D, Chen Q, Ping Y, Pang D. Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer. 2017;16(1):129. doi:10.1186/s12943-017-0696-6
29. Pawlowska E, Szczepanska J, Blasiak J. The long noncoding RNA HOTAIR in breast cancer: does autophagy play a role? Int J Mol Sci. 2017;18(11):11. doi:10.3390/ijms18112317
30. Xue X, Yang YA, Zhang A, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35(21):2746–2755. doi:10.1038/onc.2015.340
31. Nedeljkovic M, Tanic N, Prvanovic M, Milovanovic Z, Tanic N. Friend or foe: ABCG2, ABCC1 and ABCB1 expression in triple-negative breast cancer. Breast Cancer. 2021;28(3):727–736. doi:10.1007/s12282-020-01210-z
32. Liao M, Wang C, Yang B, et al. Autophagy blockade by Ai Du Qing formula promotes chemosensitivity of breast cancer stem cells via GRP78/beta-catenin/ABCG2 axis. Front Pharmacol. 2021;12:659297. doi:10.3389/fphar.2021.659297
33. Busby M, Hallett MT, Plante I. The complex subtype-dependent role of connexin 43 (GJA1) in breast cancer. Int J Mol Sci. 2018;19(3):693. doi:10.3390/ijms19030693
34. Phillips SL, Williams CB, Zambrano JN, Williams CJ, Yeh ES. Connexin 43 in the development and progression of breast cancer: what’s the connection?. Int J Oncol. 2017;51(4):1005–1013. doi:10.3892/ijo.2017.4114
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.