Back to Journals » Cancer Management and Research » Volume 11
HLA-DQB1 expression on tumor cells is a novel favorable prognostic factor for relapse in early-stage lung adenocarcinoma
Authors Zhang L , Li M, Deng B, Dai N, Feng Y, Shan J, Yang Y, Mao C, Huang P, Xu C, Wang D
Received 11 December 2018
Accepted for publication 19 February 2019
Published 1 April 2019 Volume 2019:11 Pages 2605—2616
DOI https://doi.org/10.2147/CMAR.S197855
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Liang Zhang,1 Mengxia Li,1 Bo Deng,2 Nan Dai,1 Yan Feng,1 Jinlu Shan,1 Yuxin Yang,1 Chengyi Mao,3 Ping Huang,1 Chengxiong Xu,1 Dong Wang1
1Cancer Center of Daping Hospital, Third Military Medical University, Chongqing 400042, People’s Republic of China; 2Thoracic Surgery Department of Daping Hospital, Third Military Medical University, Chongqing 400042, People’s Republic of China; 3Pathology Department of Daping Hospital, Third Military Medical University, Chongqing 400042, People’s Republic of China
Background: Postoperative recurrence is the main cause of a poor prognosis in early-stage lung adenocarcinoma (LUAD). Factors that can predict recurrence risk are critically needed.
Materials and methods: In this study, we designed a screening procedure based on gene profile data and performed validation using TCGA and Daping hospital’s cohorts. Differentially expressed genes (DEGs) between patients with recurrence-free survival (RFS) 3 years were identified, overlapping genes among these DEGs were selected as candidate biomarkers. A Cox proportional hazards model, immunohistochemistry and Kaplan-Meier survival analysis were performed to validate these biomarkers in two distinct validation sets.
Results: SFTPB, SFTPD, SFTA1P, HLA-DQB1, ITGB8, ANLN, and LRRN1 were overlapped both in TCGA and Daping discovery sets. The Cox proportional hazards model analysis of the TCGA validation set showed that HLA-DQB1 was an independent prognostic factor for RFS (HR=0.686, 95% CI, 0.542–0.868). Immunohistochemistry and Kaplan-Meier analysis in Daping validation sets confirmed HLA-DQB1 expression on tumor cells (not interstitial cells) to be an effective predictor of postoperative recurrence. Further examination revealed that the level of HLA-DQB1 expression on tumor cells was positively correlated with CD4- and CD8-positive lymphocyte infiltration into the tumor.
Conclusion: All results indicate that high expression of HLA-DQB1 on tumor cells is a good prognostic marker in early-stage LUAD, and the mechanism may be related to anti-tumor immune activity.
Keywords: HLA-DQB1, lung adenocarcinoma, tumor recurrence, prognostic factor
Introduction
Lung cancer is the leading cause of cancer incidence and mortality worldwide, with 2.1 million new lung cancer cases and 1.8 million deaths predicted in 2018.1 Approximately 60% of lung cancer patients are initially diagnosed with advanced-stage disease, and the 5-year survival rate of this subgroup is only 5% in Unite States. Among those diagnosed with early-stage disease, approximately half (30–50%) ultimately die of recurrence.2 Although adjuvant therapy is recommended for stage II and III patients,3 the benefit is limited, and only a 4% increase in the 5-year survival rate is achieved.4 In clinical practice, we have found that some patients experience recurrence within one year after curative surgery, with an extremely poor prognosis. We speculate that this group of patients may have particular biological characteristics. To maximize the benefit of adjuvant therapy, early-stage non-small cell lung cancer (NSCLC) patients should be selected based on clinical features or biomarkers before receiving this therapy. Indeed, developing biomarkers that can categorize patients according to postoperative recurrence risk is vital to improving the treatment strategy after complete resection for early-stage NSCLC.
There have been intense efforts in this field over the past decade, and a variety of predictive factors (eg, proteins,5 mRNA,6 miRNA,7 DNA methylation8) have been discovered in succession. However, heterogeneity in patient cohorts, treatment regimens, and technological platforms, among other factors, has led to apparently inconsistent results and only a modest translational impact. To address these issues, we designed a screening validation study utilizing two different lung adenocarcinoma (LUAD) cohorts (Figure 1). In this study, we integrated gene profiles derived from different platforms and different ethnicities and performed a validation procedure in two different validation sets, at both RNA and protein levels, in search for an effective predictor of postoperative recurrence in early-stage LUAD.
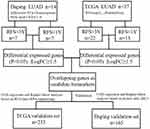 | Figure 1 Workflow of the screening and validation procedures used in this study. Abbreviations: LUAD, lung adenocarcinoma; RFS, recurrence-free survival. |
Materials and methods
Patients
The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of Daping Hospital (2017 number 30). Patients who underwent radical surgery and histologically confirmed LUAD from 2010 to 2013 at Daping Hospital were enrolled in this study. Patients who had an evidence of residual tumor at the resection margin or lymph node metastasis or dead within 30 days after surgery were excluded from this study. All patients had provided written informed consent during hospitalization. Clinical data were acquired from medical records and telephone follow-up. Tumor stage was assessed by American Joint Commission on Cancer (AJCC) 8th Edition Guidelines.9 Recurrence-free survival (RFS) was measured from the date of surgery until disease relapse or metastasis. LUAD cases with RFS<1 or >3 years (7 vs 7) were selected to construct the Daping discovery set; we then constructed the Daping validation set with 165 pathological stage I-IIIA patients.
Human transcriptome array
Transcriptome analysis was performed to identify differentially expressed genes (DEGs) between the RFS <1 year and >3 year subgroups in the Daping discovery set (Series GSE121841). Tissue cores with >70% tumor content were obtained from formalin-fixed and paraffin-embedded (FFPE) tissue. RNA was extracted from deparaffinized and proteinase K–treated FFPE tissue (20–40 mg) using RNA prep Pure FFPE Kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s instructions. The RNA concentration and quality were assessed by absorbance spectrometry using a NanoDrop (Thermo Fisher Scientific, Wilmington, DE, USA). We performed gene expression profiling of 14 FFPE-derived LUAD tissues using a GeneChip® Human Transcriptome Array 2.0 (Affymetrix, Santa Clara, CA, USA); the total RNA input was 500 pg–10 µg. Data were processed using Affymetrix® Transcriptome Analysis Console software 3.0.
TCGA data management
We obtained TCGA data for NSCLC adenocarcinoma from
Immunohistochemistry (IHC)
We followed routine methods of our laboratory for immunohistochemistry.10 LUAD FFPE tissue samples were cut into 3- to 5-µm sections and mounted on uncharged glass slides. The sections were deparaffinized, hydrated, subjected to antigen retrieval by heating in a steamer for 20 min with 10 mmol/L sodium EDTA (pH 9.0), and treated with 3% H2O2–methanol solution for 10 min to reduce endogenous peroxidase activity. The sections were then washed with PBS and incubated overnight at 4°C with a primary antibody (HLA-DQB1 GTX113458, GeneTex, Irvine, CA, USA; CD4 ZA0519 and CD8 ZA0508, ORIGENE, Beijing, CHINA). The sections were then washed and incubated with a peroxidase-conjugated anti-rabbit secondary antibody (Rabbit HRP EnVision TM+, Dako, Glostrup, Denmark) for 30 min at room temperature. After a PBS washing step, signals were developed with 3,3-diaminobenzidine (DAB) substrate for 10 min, followed by counterstaining with hematoxylin (1:4, BioGenex, Fremont, CA, USA) for 2 min. The sections were rinsed in distilled water and mounted on glass slides before evaluation under a microscope. For negative controls, FFPE samples were processed in the same manner, but the primary antibody was omitted.
Experienced pathologists blinded to the clinical data jointly examined the immunostained sections by light microscopy to generate one set of readings (Dong W and Mengxia L). HLA-DQB1 expression was estimated according to the staining intensity of cancer cells and graded as follows: negative, score 0; weak, score 1+; moderate, score 2+; or strong, score 3+. The number of CD4- and CD8-positive cells was derived using Image-Pro Plus software 6.0. We randomly selected 5 microscope fields and calculated the number of positive-staining cells and nucleated cells in each field. The percentage of CD4- or CD8-positive lymphocytes among all nucleated cells in the tumor tissue was assessed.
Statistical analysis
DEG analysis of the TCGA and Daping discovery sets was performed using R (3.32) Bioconductor Package limma and Transcriptome Analysis Console 3.0, respectively. We used four clinical variables (stage, smoking history, sex, and age) and seven candidate genes (SFTPB, SFTPD, SFTA1P, HLA-DQB1, ITGB8, ANLN, and LRRN1) to construct a Cox proportional hazard regression model for predicting prognosis. Univariate and multivariate Cox proportional hazard regression analyses were performed using SPSS 24. The Kaplan–Meier method was applied to analyze the association between HLA-DQB1 expression and survival endpoints, such as RFS. The Wilcoxon-Mann-Whitney test and χ2 test were employed to assess differences between two groups. P<0.05 indicated statistical significance.
Results
Patient characteristics
The demographics of the patients included in the Daping and TCGA discovery sets are shown in Table 1. Clinical variables (age, sex, smoking status, tumor size, adjuvant therapy, and anatomic neoplasm subdivision) were compared between the RFS <1 year and RFS >3 year groups using the χ2 test, with no statistically significant differences for any of the variables.
 | Table 1 Clinical characteristics of the Daping and TCGA discovery sets |
Table 2 summarizes the clinicopathological characteristics of the patients enrolled in Daping and TCGA validation sets. The TCGA validation set (n=233) consisted of 137 stage I and 96 stage II-IIIA LUAD patients; the Daping validation set (n=165) consisted of 131 stage I and 34 stage II-IIIA LUAD patients. Most patients in the Daping validation set (127/165) had received postoperative adjuvant therapy, whereas the status of adjuvant therapy for most of the TCGA validation set patients was unclear. Nearly half of the patients (80/165) in the Daping validation set experienced recurrence or metastasis during follow-up, and the median recurrence-free survival time was 1,975 days. Approximately 40% of the patients in the TCGA validation set suffered disease recurrence or metastasis, with a median RFS of 1,433 days. The mean follow-up time in the Daping and TCGA validation sets was 1,814 (153–3,034) and 923 (60–6,812) days, respectively.
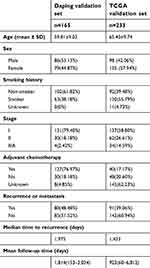 | Table 2 Clinical characteristics of the Daping and TCGA validation sets |
Candidate gene selection
As shown in Figure 1, we designed a procedure to develop and validate biomarkers for predicting recurrence risk in patients with early-stage LUAD. In step 1, we obtained lists of DEGs between patients with RFS<1 year and those with RFS>3 years through transcriptome microarray analysis of the Daping discovery set. In total, 734 genes matched the criteria (ANOVA P<0.05 and |fold change|>1.5). In step 2, we analyzed the RNA sequencing data of the TCGA discovery set using the R limma package and obtained another list containing 1,258 DEGs, with 229 matching the criteria. In step 3, overlapping genes (SFTPB, SFTPD, SFTA1P, HLA-DQB1, ITGB8, ANLN, and LRRN1) in the two DEG lists were selected as candidates (Table 3; ATP13A4 was eliminated because of inconsistent trends in expression changes). The Cox proportional hazards model analysis employed in the next validation procedure mainly focused on these genes.
 | Table 3 Overlapping genes in the Daping and TCGA DEG lists |
Validation of candidate predictors at the RNA level in the TCGA validation set
A Cox proportional hazards model was employed to determine whether these candidate genes are dependent or independent prognostic factors for RFS in early-stage LUAD. As shown in Tables 4 and 5, univariate analysis indicated that all candidate genes were associated with RFS in stage I LUAD patients; however, only HLA-DQB1, SFTPB, ANLN, SFTPD and stage reached significance for early-stage LUAD patients. Multivariate analysis confirmed only HLA-DQB1 to be an independent prognostic factor for RFS in both stage I (P=0.002, HR=0.686, 95% CI, 0.542–0.868) and early-stage (P=0.044, HR=0.875, 95% CI, 0.768–0.996) LUAD. Therefore, we conducted further validation focused on HLA-DQB1.
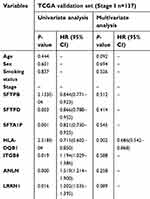 | Table 4 patients |
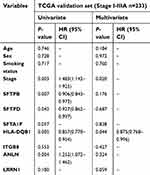 | Table 5 patients |
By evaluating RNA sequencing data of lung adenocarcinoma in the TCGA database, we found that the level of HLA-DQB1 mRNA in tumor tissues of LUAD patients was significantly lower than that in adjacent normal tissues (Wilcoxon mathced pairs test, tow-tailed t-test P<0.001) (Figure 2A). An optimal cut-off point (8,871) for HLA-DQB1 in the TCGA validation sets was determined using the X-Tile program (version 3.6.1).11 The correlation between the HLA-DQB1 mRNA level and clinical status is shown in Table 6. Patients exhibiting high expression of the HLA-DQB1 had significantly lower recurrence rates than did subgroups with low HLA-DQB1 expression (17.19% vs 47.34% χ2 test P<0.001) (Figure 2B). In addition, Kaplan-Meier survival analysis showed that HLA-DQB1 gene expression level was correlated with RFS in both stage I (P<0.0001, HR=0.2127, 95% CI, 0.1108–0.4081) and early-stage (P=0.0008, HR=0.4012, 95% CI, 0.2574–0.6255) LUAD patients (Figure 2C and D). In the TCGA validation set, the median RFS of the HLA-DQB1-high group was longer than that of the HLA-DQB1-low group (undefined vs 798 days). These results indicate that patients with lower levels of HLA-DQB1 mRNA had a higher risk of recurrence after curative surgery.
 | Table 6 Correlation between HLA-DQB1 expression and clinical characteristics in the Daping and TCGA validation sets |
Validation at the protein level in the Daping validation set
To further investigate the value of HLA-DQB1 as a prognostic factor for RFS in early-stage LUAD, IHC was performed in the Daping validation set. According to staining results, the HLA-DQB1 protein is mainly located on the cell membrane of tumor and interstitial cells (Figure S1A). Moreover, the expression level of HLA-DQB1 varied among the patients (Figure 3A). The correlation between tumor HLA-DQB1 protein expression and clinical variables is shown in Table 6. Although expression level of HLA-DQB1 in tumor cells appears to be related to sex and the pathological stage of the disease, this correlation was not observed upon TCGA verification. Approximately 60% (98/165) of patients expressed high levels of HLA-DQB1 protein (score 2 or 3) in tumor cells, and these patients had a lower recurrence rate (34% vs 68% χ2 test P<0.001) than those with lower levels (score 0 or 1) of HLA-DQB1 protein expression in tumor cells (Figure 3B).
Kaplan-Meier survival analysis also showed that patients with higher HLA-DQB1 expression in tumor cells had a longer RFS than did those with lower HLA-DQB1 expression (Figure 3C; P<0.0001), and there was a significant difference in RFS between the HLA-DQB1 high- and low-expression groups in both stage I and early-stage LUAD (Figure 3D and E). The median RFS of the low-HLA-DQB1 group was 28 months, whereas that of the high-HLA-DQB1 group was undefined. HLA-DQB1 expression in interstitial cells had no significant correlation with RFS in LUAD patients (Figure S1B).
HLA-DQB1 expression is associated with tumor immunity
HLA-DQB1 is an HLA class II beta chain paralog. Class II molecules are expressed by antigen-presenting cells and play a central role in the immune system by presenting peptides derived from extracellular proteins. It is striking that the HLA-DQB1 protein is expressed on LUAD tumor cells and associated with RFS in our study, prompting the question of whether the function of HLA-DQB1 on tumor cells is similar to that on APCs and affects antitumor immunity. To reveal the relationship between tumor-HLA-DQB1 and the antitumor immune response, the mRNA levels of immune-related genes and the lymphocyte infiltration status were first determined in the TCGA validation set. As shown in Figure 4, patients with high HLA-DQB1 levels also had a high lymphocyte infiltration rate and greater levels of CD4 and CD8 (P<0.0001). GZMK and GZMH, which mediated cytotoxic functions, were also positively correlated to HLA-DQB1. Furthermore, we found a positive correlation between the HLA-DQB1 mRNA level and the cytotoxic immune signature defined by Davoli.12
Next, we investigated the relationship between tumor HLA-DQB1 protein levels and CD4- and CD8-positive T lymphocyte infiltration status by IHC in the Daping validation set. The results were consistent with the analysis at the mRNA level in the TCGA validation set: tumors with higher HLA-DQB1 levels showed greater infiltration of CD4- and CD8-positive T lymphocytes (Figure 5).
Discussion
According to the recent National Lung Screening Trial, low-dose spiral computed tomography screening can sharply increase the number of lung cancer cases diagnosed at an early stage, and thus, more patients will be eligible for curative surgery.13,14 Accordingly, the problem of postoperative recurrence is even more important. Considering that the effect of postoperative adjuvant therapy is minimal,15,16 it is increasingly vital to develop screening biomarkers that can differentiate early-stage LUAD patients as high risk or low risk as for recurrence.
Numerous studies have identified prognostic biomarkers for NSCLC based on various technologies.5,6,8,17–21 For example, Beer D.G. utilized gene expression profiles based on microarray analysis to identify a set of genes that can predict survival in early-stage LUAD.22 Additionally, Robles and his colleague found a prognostic classifier comprising three types of genomic and epigenomic data that can help guide the postoperative treatment of stage I LUAD patients at high risk of recurrence.23 However, due to poor consistency between different studies and high costs, most gene signature predictors have not gained sufficient acceptance to become the standard of care. In our study, we integrated gene profiles derived from different platforms and different ethnicities and found tumor HLA-DQB1 levels to be associated with RFS in early-stage LUAD. It is noteworthy that nearly 80% of the patients in the Daping validation set had received adjuvant chemotherapy. Overall, Kaplan-Meier survival analysis showed that HLA-DQB1 was an effective prognostic factor regardless of whether the patient received adjuvant chemotherapy (Figure S2A and B). Furthermore, the result was validated by IHC using FFPE tissue, which is easy to perform in clinical practice and relatively inexpensive.
HLA-DQB1 is a beta chain paralog of HLA class II molecule which is critical for the immune response. Previous studies involving HLA-DQB1 have mainly focused on polymorphisms and cancer susceptibility,24,25 whereas the relationship between HLA-DQB1 expression level and tumor prognosis has rarely been reported in lung cancer. The study of He et al26 described HLA class II protein expression in lung cancer cell lines and patient tissues and found HLA class II expression by TILs was correlated with a better prognosis in NSCLC patients. In He’s study, HLA class II expression levels differed between squamous cell carcinoma and adenocarcinoma. In our study we also investigated HLA-DQB1 mRNA levels in TCGA LUSC cohorts and failed to find a correlation between HLA-DQB1 expression level and RFS in LUSC. Thus, lung squamous cell carcinoma may be a confounding factor in NSCLC research. On another hand, expression of HLA class II by TILs was examined in He’s study, whereas we only evaluated HLA-DQB1 expression by interstitial cells without using TIL markers for labeling. Nonetheless, the main results of the two studies are not conflicting.
HLA class II molecules are typically expressed on APCs and can be induced or expressed constitutively on epithelial cells. It has been reported that HLA class II expressed on epithelial cells can present antigens and activate CD4+ T cells.27,28 Sconocchia and his colleague investigated the frequency of HLA class II antigen expression in colorectal carcinoma (CRC) and found that HLA class II antigen was associated with a favorable outcome.29 However, unlike in the current study, the antibody they used was a generic antibody that can recognize HLA-DR, HLA-DQ, and HLA-DP antigens, and the overall frequency (23%) of expression in CRC was lower than that in LUAD (>50%). Regardless, the conclusion that HLA class II antigen expression on CRC tumor cells is a predictor of a favorable outcome was consistent with our study. We also found HLA-DQB1 expression on tumor cells to be a prognostic marker of overall survival (Figure S2C and D).
Moreover, some in vitro work in Sconocchia’s study revealed that HLA class II antigen expression on CRC cells can trigger interleukin-1β production and influence the antitumor immune response. In the current study, we also found that GZMK and GZMH, which mediate cytotoxic functions, were positively related to HLA-DQB1. Due to the fact that HLA class II molecules expressed by epithelial cells can present antigens and activate CD4+ T cells, the tumor antigenic potential may increase when HLA-DQB1 protein levels on tumor cells increased. Additionally, we observed a positive correlation between HLA-DQB1 and a cytotoxic immune signature, as defined by Davoli.12 In many studies, tumor-infiltrating lymphocytes (TILs), especially CD4- and CD8-positive lymphocytes, have been used as a surrogate of antitumor immune activity and as a favorable prognostic marker for survival.30,31 In our study, we found that HLA-DQB1 expression positively correlated with CD4- and CD8-positive lymphocyte infiltration in LUAD tumors. It is further demonstrated that HLA-DQB1 may be associated with anti-tumor immunity.
In conclusion, HLA-DQB1 protein expression on tumor cells is a robust predictive biomarker for RFS in early-stage LUAD, and the mechanism may be related to antitumor immunity. We speculate that early-stage LUAD tumors with high HLA-DQB1 protein expression may present high antigenic potential and trigger the immune system, especially T cells, with more efficient elimination of residual tumor cells after curative resection. In contrast, tumors with low HLA-DQB1 protein expression may easily escape immune destruction and ultimately recur. Antitumor immunity is complex, and further studies are needed to uncover the relationship with HLA-DQB1 expression. Prospective studies are also needed to test the clinical utility of HLA-DQB1.
Acknowledgments
The authors thank Quan-Zhen Li, M.D, Ph.D. and the members of his teams at UT Southwestern Medical Center. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Disclosure
The authors report no conflicts of interest in this study.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
3. Ettinger DS, Wood DE, Aisner DL, et al. Non-small cell lung cancer, version 5.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2017;15(4):504–535.
4.
5. Gold KA, Kim ES, Liu DD, et al. Prediction of survival in resected non-small cell lung cancer using a protein expression-based risk model: implications for personalized chemoprevention and therapy. Clin Cancer Res. 2014;20(7):1946–1954. doi:10.1158/1078-0432.CCR-13-1959
6. Wistuba II, Behrens C, Lombardi F, et al. Validation of a proliferation-based expression signature as prognostic marker in early stage lung adenocarcinoma. Clin Cancer Res. 2013;19(22):6261–6271. doi:10.1158/1078-0432.CCR-13-0596
7. Lu Y, Govindan R, Wang L, et al. MicroRNA profiling and prediction of recurrence/relapse-free survival in stage I lung cancer. Carcinogenesis. 2012;33(5):1046–1054. doi:10.1093/carcin/bgs100
8. Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358(11):1118–1128. doi:10.1056/NEJMoa0706550
9. Amin MB, Greene FL, Edge SB, et al. The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin. 2017;67:93–99. doi:10.3322/caac.21388
10. Li Z, Qing Y, Guan W, et al. Predictive value of APE1, BRCA1, ERCC1 and TUBB3 expression in patients with advanced non-small cell lung cancer (NSCLC) receiving first-line platinum-paclitaxel chemotherapy. Cancer Chemother Pharmacol. 2014;74(4):777–786. doi:10.1007/s00280-014-2562-1
11. Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–7259. doi:10.1158/1078-0432.CCR-04-0713
12. Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355.
13.
14. van der Aalst CM, Ten Haaf K, de Koning HJ. Lung cancer screening: latest developments and unanswered questions. Lancet Respir Med. 2016;4(9):749–761. doi:10.1016/S2213-2600(16)30200-4
15. West H. Individualizing adjuvant therapy for early stage non-small cell lung cancer: we see the destination, but we don’t yet know the route. J Thorac Dis. 2015;7(3):235–237. doi:10.3978/j.issn.2072-1439.2015.01.08
16. Pakkala S, Ramalingam SS. Adjuvant therapy for nonsmall cell lung cancer: recent advances and future perspectives. Curr Opin Oncol. 2016;28(2):150–158. doi:10.1097/CCO.0000000000000269
17. Lee ES, Son DS, Kim SH, et al. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin Cancer Res. 2008;14(22):7397–7404. doi:10.1158/1078-0432.CCR-07-4937
18. Chen HY, Yu SL, Chen CH, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356(1):11–20. doi:10.1056/NEJMoa060096
19. Raponi M, Zhang Y, Yu J, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66(15):7466–7472. doi:10.1158/0008-5472.CAN-06-1191
20. Lu Y, Lemon W, Liu PY, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med. 2006;3(12):e467. doi:10.1371/journal.pmed.0030467
21. Okayama H, Schetter AJ, Ishigame T, et al. The expression of four genes as a prognostic classifier for stage I lung adenocarcinoma in 12 independent cohorts. Cancer Epidemiol Biomarkers Prev. 2014;23(12):2884–2894. doi:10.1158/1055-9965.EPI-14-0182
22. Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8(8):816–824. doi:10.1038/nm733
23. Robles AI, Arai E, Mathe EA, et al. An integrated prognostic classifier for stage I lung adenocarcinoma based on mRNA, microRNA, and DNA methylation biomarkers. J Thorac Oncol. 2015;10(7):1037–1048. doi:10.1097/JTO.0000000000000560
24. Wank R, Meulen JT, Luande J, Eberhardt HC, Pawlita M. Cervical intraepithelial neoplasia, cervical carcinoma, and risk for patients with HLA-DQB1*0602,*301,*0303 alleles. Lancet. 1993;341(8854):1215. doi:10.1016/0140-6736(93)91043-L
25. David AL, Taylor GM, Gokhale D, et al. HLA-DQB1*03 and cervical intraepithelial neoplasia type III. Lancet. 1992;340(8810):52. doi:10.1016/0140-6736(92)92464-Q
26. He Y, Rozeboom L, Rivard CJ, et al. MHC class II expression in lung cancer. Lung Cancer. 2017;112:75–80. doi:10.1016/j.lungcan.2017.07.030
27. Londei M, Lamb JR, Bottazzo GF, Feldmann M. Epithelial cells expressing aberrant MHC class II determinants can present antigen to cloned human T cells. Nature. 1984;312(5995):639–641.
28. Herkel J, Jagemann B, Wiegard C, et al. MHC class II-expressing hepatocytes function as antigen-presenting cells and activate specific CD4 T lymphocyutes. Hepatology. 2003;37(5):1079–1085. doi:10.1053/jhep.2003.50191
29. Sconocchia G, Eppenberger-Castori S, Zlobec I, et al. HLA class II antigen expression in colorectal carcinoma tumors as a favorable prognostic marker. Neoplasia. 2014;16(1):31–42.
30. Al-Shibli KI, Donnem T, Al-Saad S, Persson M, Bremnes RM, Busund L-T. Prognostic effect of epithelial and stromal lymphocyte infiltration in non-small cell lung cancer. Clin Cancer Res. 2008;14(16):5220–5227. doi:10.1158/1078-0432.CCR-08-0133
31. Brambilla E, Le Teuff G, Marguet S, et al. Prognostic effect of tumor lymphocytic infiltration in resectable non-small-cell lung cancer. J Clin Oncol. 2016;34(11):1223–1230. doi:10.1200/JCO.2015.63.0970
Supplementary materials
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.