Back to Journals » Infection and Drug Resistance » Volume 13
HIV-1 Drug Resistance in ART-Naïve Individuals in Myanmar
Authors Ye M, Chen X, Wang Y, Zhou YH, Pang W, Zhang C, Zheng YT
Received 17 January 2020
Accepted for publication 31 March 2020
Published 20 April 2020 Volume 2020:13 Pages 1123—1132
DOI https://doi.org/10.2147/IDR.S246462
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Mei Ye,1,2 Xin Chen,1,3 Yu Wang,4 Yan-Heng Zhou,5 Wei Pang,1 Chiyu Zhang,6 Yong-Tang Zheng1,4
1Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences/Key Laboratory of Bioactive Peptides of Yunnan Province, KIZ-CUHK Joint Laboratory of Bioresources and Molecular Research in Common Diseases, Center for Biosafety Mega-Science, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, People’s Republic of China; 2Savaid Medical School, University of Chinese Academy of Sciences, Beijing 101408, People’s Republic of China; 3Department of Pathogenic Biology, School of Basic Medical Sciences, Gannan Medical University, Ganzhou 341000, People’s Republic of China; 4KIZ-SU Joint Laboratory of Animal Model and Drug Development, College of Pharmaceutical Sciences, Soochow University, Suzhou, Jiangsu 215000, People’s Republic of China; 5Shaanxi Engineering and Technological Research Center for Conversation and Utilization of Regional Biological Resources, College of Life Sciences, Yan’an University, Yan’an, Shaanxi 716000, People’s Republic of China; 6Pathogen Discovery and Evolution Unit, Institute Pasteur of Shanghai, Chinese Academy of Sciences, Shanghai 200025, People’s Republic of China
Correspondence: Yong-Tang Zheng
Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences, Kunming Institute of Zoology, Chinese Academy of Sciences, 32 Jiaochang Donglu, Kunming, People’s Republic of China
Tel/Fax +86 871 65195684
Email [email protected]
Chiyu Zhang
Pathogen Discovery and Evolution Unit, Institute Pasteur of Shanghai, 320 Yueyang Road, Shanghai, People’s Republic of China
Tel/Fax +86 21 54923051
Email [email protected]
Background: Estimating the prevalence and characterizing the transmission of HIV-1 drug resistance in treatment-naïve individuals are very important in the prevention and control of HIV/AIDS. As one of the areas most affected by HIV/AIDS, few data are currently available for HIV-1 drug resistance in antiretroviral therapy (ART)-naïve individuals in Myanmar, which borders Yunnan, China.
Methods: HIV-1 pol sequences from ART-naïve HIV-1-infected individuals during 2008 and 2014 in Myanmar were retrieved from our previous studies. HIV-1 transmitted drug resistance (TDR) and susceptibility to antiretroviral drugs were predicted using the Stanford HIVdb program. HIV-1 transmission cluster (TC) was determined by Cluster Picker.
Results: A total of 169 partial pol sequences from ART-naïve HIV-1 positive Burmese were analyzed. The prevalence of TDR was 20.1%. CRF01_AE and BC recombinants appeared to have a higher prevalence of TDR than other subtypes. The V179D/T was found to be very common in the China–Myanmar border region and was involved in half of the transmission clusters formed by HIV-1 drug-resistance strains in this region. Comparison showed that drug-resistance mutation profile in Myanmar was very similar to that in Dehong prefecture of Yunnan. By further phylogenetic analysis with all available sequences from the China–Myanmar border region, four HIV-1 drug-resistance-related TCs were identified. Three of them were formed by Burmese long-distance truck drivers and the Burmese staying in Yunnan, and another was formed by Burmese injection drug users staying in Myanmar and Yunnan. These results suggest a potential transmission link of HIV-1 drug resistance between Myanmar and Yunnan.
Conclusion: Given the high prevalence of TDR in Myanmar, and the potential risk of cross-border transmission of HIV-1 drug-resistant strains between Myanmar and Yunnan, China, ongoing monitoring of HIV-1 drug resistance in ART-naïve individuals will provide a guideline for clinical antiretroviral treatment and benefit the prevention and control of HIV/AIDS in this border region.
Keywords: HIV-1, antiretroviral therapy, transmission cluster, drug-resistance mutations, ART-naïve, Myanmar, Yunnan
Introduction
According to the World Health Organization (WHO), 23.3 million people living with HIV (PLHIV) had accessed antiretroviral therapy (ART) by the end of 2018 globally.1 The global scale-up of ART has led to a significant decline in HIV-1 related mortality and morbidity.2–4 However, the emergence and spread of HIV-1 drug resistance are major challenges to the long-term success of ART and have important clinical significance and public health implication including increasing risks of first-line ART failure and onward transmission of drug-resistance strains in ART-naïve individuals.5–8 In particular, transmitted drug resistance (TDR) or pre-treatment drug resistance (PDR) can increase the risk of virological failure (VF), and compromise the long-term effectiveness of recommended first-line ART regimens after initiating ART.9
HIV-1 strains with transmission links can form genetic transmission clusters, which are especially helpful for tracing the transmission source. Intervention for individuals within a large cluster by prioritizing ART is an effective strategy to reduce new infections.10 In recent years, transmission clusters formed by TDR were reported among the ART-naïve individuals in some countries, such as China,11 Denmark,12 Germany,13 Switzerland,14 Britain,15 Greece,5 Italy,16 Spain,17 America,18 and Canada,19 implying the importance of TDR monitoring. Whether the ART rollout in resource-limited settings also drives the formation of transmission clusters of TDR deserves investigation.
Myanmar is a lower-middle-income country with limited resources, located near the “Golden Triangle”, a well-known region producing illicit drugs in Southeast Asia.20,21 Injection drug use has been a main risk factor for HIV-1 epidemic in Myanmar over the past three decades.22 The number of estimated PLHIV in Myanmar was 240,000 in 2018, and 7800 people died from AIDS-related diseases.23 Although the coverage of ART raised about three times from 24% in 2012 to 70% in 2018, Myanmar still faces a heavy burden of HIV/AIDS with 11,000 new infections reported in 2018.23 A recent study focusing on Burmese adolescents and adults receiving first-line ART during 2005 to 2015 reported a high rate (3.2 per 100 person-years follow-up) of virological failure but a low opportunity (1.4 per 100 person-years follow-up) to switch to second-line ART regimens.24 High prevalence of drug resistance among ART-treated individuals can spread HIV-1 drug resistance to ART-naïve individuals, and lead to a high proportion of VF in the latter after they initiate ART.9,25,26
Northern Myanmar borders Yunnan, China, and is the hardest-hit region by the HIV-1 epidemic.27 Our previous studies showed that northern Myanmar had a high prevalence of HIV-1 recombinants and demonstrated that Burmese long-distance truck drivers (LDTDs) and injection drug users (IDUs) staying in Yunnan could form transmission clusters with local IDUs and contribute to the cross-border transmission of HIV-1 and other viruses between Myanmar and China.28–31 In particular, a high prevalence (12.8%) of HIV-1 TDR was detected among ART-naïve Burmese travelers entering Yunnan, China via Dehong ports during 2003 to 2012,32 implying a cross-border transmission risk of HIV-1 drug resistance.13 Whether the cross-border activities of these high-risk Burmese groups of IDUs and LDTDs drive the cross-border transmission of HIV-1 drug resistance via transmission links with local counterparts needs to be determined.
In the present study, we investigated HIV-1 drug resistance among ART-naïve Burmese residing in Myanmar by a retrospective analysis of previously reported HIV-1 pol sequences and evaluated the cross-border transmission risk of TDR by phylogenetic analysis of available pol sequences from ART-naïve individuals in the China–Myanmar border region.
Materials and Methods
Sequence Data Collection
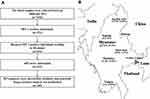
This retrospective study is based on data from our surveys done in the China–Myanmar border region from 2008 to 2014. To investigate the prevalence of HIV-1 drug resistance among ART-naïve Burmese, HIV-1 pol sequences were retrieved and downloaded from the HIV-1 Sequence Database (www.hiv.lanl.gov) according to our four previous studies.28,30,33,34 This study is a second analysis of these sequence data. The study participants should meet the following inclusion criteria: (i) were Burmese and resided in Myanmar; (ii) did not accept antiretroviral drugs (Figure 1A). The protocol of this study was approved by the Ethics Committee of Kunming Institute of Zoology, Chinese Academy of Sciences (approval number: SWYX-2008010; SWYX-2012017). All the participants were provided written informed consent.
Sequence Processing and Phylogenetic Analysis
The qualified pol sequences were aligned with reference sequences using MEGA 7 and then manually edited. A fragment of the HIV-1 partial pol gene encompassing the whole protease and 267 codons of the reverse transcriptase (HXB2: 2253–3350) were obtained after realigning partial pol (HXB2: 2147–3462) and near full-length HIV-1 genome (HXB2: 809–9124) and trimming them equivalent length.
The maximum likelihood (ML) phylogenetic trees were estimated using the general time-reversible nucleotide substitution model with gamma rate heterogeneity among sites and 1000 bootstrap replicates were implemented in MEGA version 7.0. Transmission clusters (TCs) of HIV-1 sequences were extracted from the ML tree using Cluster Picker software. Transmission clusters were defined as those clusters with ≥2 sequences having node support threshold greater than 90% and within-cluster genetic distance less than 3.0% nucleotide substitutions per site according to the previous articles.35,36 A large cluster was defined as having more than 10 isolates. Phylogenetic trees were visualized using software FigTree v1.4.3.
Genotypic Drug-Resistance Analysis
To identify surveillance drug-resistance mutations (SDRMs) and estimate the level of HIV-1 drug resistance to antiretroviral drugs, eligible HIV-1 partial pol sequences (Protease codons: 1–99; Reverse Transcriptase codons: 1–267) were analyzed using an online tool in the Stanford HIV-1 Drug Resistance Database (http://hivdb.stanford.edu). The Stanford algorithm classifies HIV-1 drug resistance into five levels: susceptible, potential low-level, low-level, intermediate and high-level drug resistance, while sequences classified as the latter three types are considered as drug resistance with a clinical impact.
Cross-Border Transmission Analysis
To explore whether there was a cross-border transmission of the HIV-1 resistant virus among ART-naïve individuals of the China–Myanmar border region, the drug-resistance mutation sequences among ART-naïve individuals from Dehong, Yunnan province, China were downloaded.29,32,37,38 After aligning with reference sequences and resistance sequences generated in this study, ML phylogenetic analysis and transmission cluster analysis were performed as described above.
Results
Socio-Demographic Characteristics of Study Population
A total of 7454 individuals were recruited in this study from 2008 to 2014. Of them, 373, including 268 IDUs and 105 LDTDs, were HIV-1 seropositive. HIV-1-positive individuals had a median age of 32 years old and came from Myanmar (n=234) and Dehong, Yunnan (n=139) (Table 1; Figure 1A). Most of these individuals were male (97.9%) and farmers (45.6%) (Table 1). There were 219 Burmese ART-naïve HIV-1 positive individuals who resided in Myanmar. Among ART-naïve individuals, 169 partial pol sequences were successfully retrieved (Table 1; Figure 1A), including 78 from Kachin State, 49 from Mandalay, 35 from Shan State, 4 from Yangon and 2 from Sagaing. Only one individual did not report the exact living address in Myanmar (Table 1; Figure 1B).
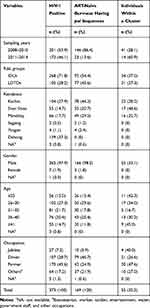 |
Table 1 Social-Demographic Characteristics of Study Subjects |
HIV-1 Drug Resistance Among ART-Naïve Burmese
Of 169 sequences, 48 (28.4%), 25 (14.8%), 11 (6.5%), 9 (5.3%), 4 (2.4%), 3 (1.8%) and 1 (0.6%) belong to CRF01_AE, C, CRF83_cpx, B, CRF82_cpx, CRF08_BC, and CRF96_cpx, respectively (Figures 2 and 3A). In particular, there are 68 sequences (40.2%) belonging to new HIV-1 recombinants, including 55 (32.5%) RFs_BC and 13 (7.7%) CRF01_AE-related RFs (Figures 2 and 3A). 34 sequences were identified to carry at least one SDRM, giving a TDR prevalence of 20.1% (Figures 1B and 2). The prevalence of HIV-1 drug resistance appeared to vary largely in different subtypes. Of 34 sequences with any SDRM, 15 and 12 belonged to CRF01_AE and RFs_BC, corresponding to prevalence of 31.2% and 21.8%, respectively (Figure 3A). The percentage of TDR was 21.8%, 14.3%, 22.4%, 0%, and 25.0% in Kachin state, Shan state, Mandalay, Sagaing and Yangon, respectively (Figure 1B).
In 34 sequences with SDRMs, 5, 29, and 7 were associated with the resistance to nucleoside reverse transcriptase inhibitors (NRTIs), non-nucleoside reverse transcriptase inhibitors (NNRTIs), and protease inhibitors (PIs), respectively (Figures 2 and 3B). The most frequently observed SDRMs were V179D/T (8.3%), E138A (4.1%) and L33F (2.4%), which are involved in the resistance to NNRTIs, NNRTIs, and PIs, respectively (Figures 2 and 3B). Of note, six individuals (6/169, 3.6%) were found harboring multiple SDRMs (Figure 2).
Among 169 ART-naïve individuals, 14 (8.3%) individuals were predicted to have clinically significant resistance to 12 antiretroviral drugs (Figures 2 and 3C). NNRTI-associated resistance was most common (7.7%, 13/169), followed by PI-associated (1.2%, 2/169) and NRTI-associated (1.2%, 2/169) resistances. Two individuals (KC913744 and KC913833) showed dual-resistance to both NRTI and NNRTI drugs, and one individual (KC913791) showed dual-resistance to both PI and NNRTI drugs (Figure 2). Four individuals (28.6%, 4/14) were predicted to have high-level resistance to NRTI and/or NNRTI drugs. There was no sequence showing high-level resistance to PIs.
HIV-1 Transmission Clusters Among ART-Naïve Burmese
There were 19 TCs having been identified, covering 55 sequences (32.5%) from ART-naïve Burmese. Ten TCs were formed by IDUs and nine TCs were formed by LDTDs (Table 1; Figure 2). The largest TC was observed with 11 CRF83_cpx sequences isolated from IDUs in Shan State. Among 19 TCs, three (clusters 5, 10 and 14) contained at least two drug-resistant strains carrying a single identical SDRM (Figure 2). Two CRF01_AE strains in the TC 10 carried two SDRMs -A62V and V179T. These results strongly suggest that the drug-resistant strains were transmitted. Furthermore, some SDRM-carrying strains were dispensed in the strains without SDRMs (Figure 2), implying that they were more likely generated by natural polymorphism of HIV-1.
Transmission Cluster Characteristics of the Drug-Resistant Strains in the China–Myanmar Border Region
To further assess the transmission of HIV-1 drug-resistant strains in ART-naïve individuals in the China–Myanmar border region, phylogenetic and transmission cluster analyses were conducted. Of all 941 sequences available from the previous studies29,32,37,38 and this study, 171 (18.2%) had at least one SDRM. Overall, 151 (16.0%) had NNRTI mutations, 15 (1.6%) had NRTI mutations, and 21 (2.2%) had PI mutations. Of those with SDRMs, 156 (91.2%) had single class resistance, 14 (8.2%) had dual-class resistance, and one (0.6%) had triple class resistance. The most frequently observed SDRM was NNRTI mutation V179D/T, which appeared in 86 participants.
Phylogenetic analysis disclosed 12 TCs involving 26 (15.2%) sequences with any SDRM (Figure 4). The majority (83.3%) of the TCs contained two individuals. Two TCs (Clusters 3 and 8) contained three individuals who carried single SDRM E138A or A98G. There were four TCs involving V179T-carrying strains, and five TCs for L33F-, L90M-, A98G-, E138A- and T215TADN-carrying strains each. Three TCs were formed by strains carrying two SDRMs, including L33F/V179T-carrying strains, A62V/V179T-carrying strains and A98G/Y181C-carrying strains (Figure 4). Importantly, we found that the V179T-carrying variants appeared in half (50%) of identified TCs.
 |
Figure 4 Maximum likelihood tree of the drug-resistance sequences among ART-naïve individuals in the China–Myanmar border region. The sequences from the Burmese IDUs,28,33,34 Burmese LDTDs,30 the Burmese staying in Yunnan29,32 and newly diagnosed HIV-1 infections in Dehong, Yunnan province37,38 are highlighted by green spots, green triangles, green diamonds and red stars, respectively. Mutations associated with resistance to PI, NRTI and NNRTI are indicated with magenta, red and blue, respectively. The brackets indicate transmission clusters that were identified by Cluster Picker. |
On the other hand, we found that all TCs were formed by Burmese individuals or Chinese individuals. Although there was no TC containing both Burmese and Chinese individuals, three TCs were found containing Burmese LDTD and the Burmese staying in Yunnan, and one TC was found containing two Burmese IDUs with one staying in Myanmar and one in Yunnan.
Discussion
In this study, we conducted a retrospective survey about HIV-1 drug-resistance characteristics in ART-naïve individuals in Myanmar and investigated the transmission link of HIV-1 drug-resistant strains in both Myanmar and Yunnan province of China. We found the prevalence of TDR was 20.1% in Myanmar. The NNRTI mutation V179D/T was very common in the China–Myanmar border region and appeared in half of the transmission clusters formed by HIV-1 drug-resistance strains in this region. These findings provide insights into the treatment and management of AIDS patients in the border region between Myanmar and China.
Myanmar has a high burden of HIV-1 with a 0.8% HIV-1 prevalence, the second highest prevalence of HIV-1 in Southeast Asia.23 Currently, the first-line ART regimen containing 2 NRTIs plus 1 NNRTI or 1 integrase inhibitor is recommended for adults and adolescents in Myanmar.39 As of 30 June, 2019, 175,297 PLHIV in Myanmar were receiving antiretroviral treatment.23 The scale-up of antiretroviral treatment inevitably increased the risk of VF and the emergence of drug resistance. Few data were available for the prevalence of HIV-1 drug resistance in ART-naïve individuals in Myanmar. Recently, WHO reported a PDR prevalence of 5.4% in Myanmar.40 Some SDRMs, including NRTI-related mutations (M41L, D67E/G/N, K70E/R, M184V, T215any, and K219any) and NNRTI-related mutations (K101E/P, K103N/S, V106A/M, Y181any, G190any, and P225H) were reported in Myanmar.40 Another survey focusing on Burmese individuals staying in Dehong reported that M184V, K103N/KN, and T74S/ST were the major SDRMs associated with VF.41 In this study, the TDR prevalence was estimated to be 20.1% in Myanmar, higher than the previously reported level,32 suggesting that drug resistance in Myanmar should not be overlooked. Furthermore, we found that the major NNRTI-related SDRMs were K101P, K103N, V106A, E138A, Y181C, and Y188H, and the major NRTI-related SDRMs were L74V/I and M184V. It was not surprising that the most common SDRMs identified in ART-naïve individuals were consistent with those identified in ART-treated individuals, suggesting a potential transmission link from patients failing treatment with resistant strains to ART-naïve individuals.11
On the other hand, the drug-resistance characteristics in Myanmar were similar to those in Dehong prefecture of Yunnan, where the NNRTI-related SDRMs dominated HIV-1 drug resistance (Figures 2 and 4). Myanmar shares border with India, China, Bangladesh, Laos and Thailand. Similar SDRM profiles were also observed among ART-naïve HIV-1 individuals in Thailand,42 India,43 and China,11,37 suggesting a possibility of cross-border transmission of HIV-1 drug resistance. This hypothesis was supported by a previous observation that TDRs circulating in the wives of IDUs in Manipur, India, were introduced from Myanmar, Vietnam and/or mainland India.43
Our previous studies reported a bidirectional cross-border transmission of HIV-1 between Myanmar and China and demonstrated that Burmese LDTDs and Burmese IDUs staying in Yunnan played a crucial role in this cross-border transmission.29,30,44 In this study, we identified some transmission clusters formed by HIV-1 drug-resistant strains in the China–Myanmar border region. Although Burmese and Chinese individuals were less inclined to share some high-risk behaviors (eg, IDU) each other,31 Burmese individuals staying in Yunnan were significantly associated with the dissemination of HIV-1 across the China–Myanmar border region.29,45 The finding that some TCs of HIV-1 drug resistance were formed by Burmese individuals staying in Myanmar and Yunnan, China, further highlighted the bridge role of Burmese individuals staying in Yunnan in the dissemination of HIV-drug-resistance strains across the China–Myanmar border region. Yunnan government is enlarging ART coverage for Burmese individuals staying in Yunnan.45 The increased cross-border transmission risk of HIV-1 drug-resistant strains accompanied by the growing number of migrant Burmese population year by year will inevitably increase the burden of monitoring HIV-1 drug resistance and managing AIDS patients for both sides of the border.
A large number of evidences showed that HIV-1 drug resistance was correlated with HIV-1 subtypes.11,46-48 The China–Myanmar border region is a hot-spot region for various HIV-1 inter-subtype recombinants formed by CRF01_AE, B and C, and has a very complex HIV-1 genetic diversity.28,33,37,38 A high proportion of drug-resistance mutations was found in ART-naïve individuals infected with HIV-1 CRF01_AE and BC recombinants with V179D/T and E138A being the most commonly observed SDRMs. Although V179D/T was more frequently transmitted in ART-naïve Burmese individuals, the presence of V179D/T did not intervene with the response to a first-line efavirenz (EFV) containing regimen.49 E138A confers low-level resistance to rilpivirine (RPV) and is more likely a consequence of HIV-1 natural polymorphism.47 Currently, tenofovir (TDF), lamivudine (3TC)/emtricitabine (FTC) and efavirenz (EFV) is the preferentially and frequently used ART first-line regime for initiation of ART in ART-naïve individuals in Myanmar and the patients receiving TDF+3TC+EFV regimen had better adherence and were less likely to switch to a protease inhibitor-containing second-line ART regimen.24,39 Therefore, increasing the coverage of the first-line regimen will benefit most ART-naïve individuals, and contribute to the control of TDRs. In spite of this, in view of high HIV-1 genetic diversity and high prevalence of TDRs in ART-naïve individuals in the China–Myanmar border region, the molecular monitoring of HIV-1 drug resistance should be strengthened.
This is a retrospective study that covers samples from 2008 to 2014. Because the sample size was relatively small and the latest samples were collected in 2014, these results may not represent the current status of HIV-1 drug resistance in ART-naïve individuals in Myanmar. In addition, relatively few sequences were available for the analyses, which might lead to an underestimate in the number of sequences involving in transmission clusters. Furthermore, it also limits our ability to assess the dynamic changes of HIV-1 drug resistance and SDRMs in ART-naïve individuals in the China–Myanmar border region. Despite these limitations, this study presents basic data on HIV-1 drug resistance in ART-naïve individuals in Myanmar, which will provide valuable policy guidance for the management and treatment of HIV/AIDS in Myanmar.
Conclusion
We report an overall HIV-1 TDR prevalence of 20.1% in Myanmar and characterize the SDRM profile. The cross-border transmission of HIV-1 drug resistance was observed in the China–Myanmar border region, and ART-naïve Burmese individuals staying in Yunnan might contribute to this transmission. The findings will benefit Myanmar, as well as China, in the prevention and control of HIV/AIDS.
Acknowledgments
We acknowledge all the participants who participated in this research. This work was supported by grants from the National Natural Science Foundation of China (U1302224, 81271892, 81601802). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors declare that they have no competing interests.
References
1. World Health Organization [homepage on the Internet]. HIV/AIDS data and statistics; 2019. Available from: https://www.who.int/hiv/data/en/.
2. Trickey A, May MT, Vehreschild JJ, et al. Survival of HIV-positive patients starting antiretroviral therapy between 1996 and 2013: a collaborative analysis of cohort studies. Lancet HIV. 2017;4(8):e349–e356. doi:10.1016/S2352-3018(17)30066-8
3. Eaton JW, Johnson LF, Salomon JA, et al. HIV treatment as prevention: systematic comparison of mathematical models of the potential impact of antiretroviral therapy on HIV incidence in South Africa. PLoS Med. 2012;9(7):e1001245. doi:10.1371/journal.pmed.1001245
4. Murphy EL, Collier AC, Kalish LA, et al. Highly active antiretroviral therapy decreases mortality and morbidity in patients with advanced HIV disease. Ann Intern Med. 2001;135(1):17–26. doi:10.7326/0003-4819-135-1-200107030-00005
5. Paraskevis D, Kostaki E, Magiorkinis G, et al. Prevalence of drug resistance among HIV-1 treatment-naive patients in Greece during 2003–2015: transmitted drug resistance is due to onward transmissions. Infect Genet Evol. 2017;54:183–191. doi:10.1016/j.meegid.2017.07.003
6. Stadeli KM, Richman DD. Rates of emergence of HIV drug resistance in resource-limited settings: a systematic review. Antivir Ther. 2013;18(1):115–123. doi:10.3851/IMP2437
7. Socias ME, Nosova E, Kerr T, et al. Patterns of transmitted drug resistance and virological response to first-line antiretroviral treatment among Human Immunodeficiency virus-infected people who use illicit drugs in a Canadian setting. Clin Infect Dis. 2017;65(5):796–802. doi:10.1093/cid/cix428
8. Stekler JD, Milne R, Payant R, et al. Transmission of HIV-1 drug resistance mutations within partner-pairs: a cross-sectional study of a primary HIV infection cohort. PLoS Med. 2018;15(3):e1002537. doi:10.1371/journal.pmed.1002537
9. Clutter DS, Jordan MR, Bertagnolio S, Shafer RW. HIV-1 drug resistance and resistance testing. Infect Genet Evol. 2016;46:292–307. doi:10.1016/j.meegid.2016.08.031
10. Smith DM, May SJ, Tweeten S, et al. A public health model for the molecular surveillance of HIV transmission in San Diego, California. AIDS. 2009;23(2):225–232. doi:10.1097/QAD.0b013e32831d2a81
11. Zuo L, Liu K, Liu H, et al. Trend of HIV-1 drug resistance in China: a systematic review and meta-analysis of data accumulated over 17 years (2001–2017). EClinicalMedicine. 2020;18:100238. doi:10.1016/j.eclinm.2019.100238
12. Petersen A, Cowan SA, Nielsen J, Fischer TK, Fonager J. Characterisation of HIV-1 transmission clusters and drug-resistant mutations in Denmark, 2004 to 2016. Euro Surveill. 2018;23(44). doi:10.2807/1560-7917.ES.2018.23.44.1700633
13. Stecher M, Chaillon A, Eis-Hubinger AM, et al. Pretreatment human immunodeficiency virus type 1 (HIV-1) drug resistance in transmission clusters of the Cologne-Bonn region, Germany. Clin Microbiol Infect. 2019;25(2):253. doi:10.1016/j.cmi.2018.09.025
14. Drescher SM, von Wyl V, Yang WL, et al. Treatment-naive individuals are the major source of transmitted HIV-1 drug resistance in men who have sex with men in the Swiss HIV Cohort Study. Clin Infect Dis. 2014;58(2):285–294. doi:10.1093/cid/cit694
15. Mbisa JL, Fearnhill E, Dunn DT, Pillay D, Asboe D, Cane PA. Evidence of self-sustaining drug resistant HIV-1 lineages among untreated patients in the United Kingdom. Clin Infect Dis. 2015;61(5):829–836. doi:10.1093/cid/civ393
16. Fabeni L, Alteri C, Di Carlo D, et al. Dynamics and phylogenetic relationships of HIV-1 transmitted drug resistance according to subtype in Italy over the years 2000–14. J Antimicrob Chemother. 2017;72(10):2837–2845. doi:10.1093/jac/dkx231
17. Vega Y, Delgado E, Fernandez-Garcia A, et al. Epidemiological surveillance of HIV-1 transmitted drug resistance in Spain in 2004–2012: relevance of transmission clusters in the propagation of resistance mutations. PLoS One. 2015;10(5):e0125699. doi:10.1371/journal.pone.0125699
18. Jair K, McCann CD, Reed H, et al. Validation of publicly-available software used in analyzing NGS data for HIV-1 drug resistance mutations and transmission networks in a Washington, DC, Cohort. PLoS One. 2019;14(4):e0214820. doi:10.1371/journal.pone.0214820
19. Brenner BG, Roger M, Moisi DD, et al. Transmission networks of drug resistance acquired in primary/early stage HIV infection. AIDS. 2008;22(18):2509–2515. doi:10.1097/QAD.0b013e3283121c90
20. The World Bank [homepage on the Internet]. The World Bank in Myanmar; 2019. Available from: https://www.worldbank.org/en/country/myanmar/overview.
21. Kusagawa S, Sato H, Watanabe S, et al. Genetic and serologic characterization of HIV type 1 prevailing in Myanmar (Burma). AIDS Res Hum Retroviruses. 1998;14(15):1379–1385. doi:10.1089/aid.1998.14.1379
22. Htoon MT, Lwin HH, San KO, Zan E, Thwe M. HIV/AIDS in Myanmar. AIDS. 1994;8(Suppl 2):S105–S109.
23. The Joint United Nations Programme on HIV/AIDS [homepage on the Internet]. HIV and AIDS estimates; 2019. Available from: http://aidsinfo.unaids.org/.
24. Kyaw NT, Harries AD, Kumar AM, et al. High rate of virological failure and low rate of switching to second-line treatment among adolescents and adults living with HIV on first-line ART in Myanmar, 2005–2015. PLoS One. 2017;12(2):e0171780. doi:10.1371/journal.pone.0171780
25. Rhee SY, Blanco JL, Jordan MR, et al. Geographic and temporal trends in the molecular epidemiology and genetic mechanisms of transmitted HIV-1 drug resistance: an individual-patient- and sequence-level meta-analysis. PLoS Med. 2015;12(4):e1001810. doi:10.1371/journal.pmed.1001810
26. Zazzi M, Hu H, Prosperi M. The global burden of HIV-1 drug resistance in the past 20 years. PeerJ. 2018;6:e4848. doi:10.7717/peerj.4848
27. Zhou YH, Liu FL, Yao ZH, et al. Comparison of HIV-, HBV-, HCV- and co-infection prevalence between Chinese and Burmese intravenous drug users of the China-Myanmar border region. PLoS One. 2011;6(1):e16349. doi:10.1371/journal.pone.0016349
28. Pang W, Zhang C, Duo L, et al. Extensive and complex HIV-1 recombination between B’, C and CRF01_AE among IDUs in south-east Asia. AIDS. 2012;26(9):1121–1129. doi:10.1097/QAD.0b013e3283522c97
29. Chen X, Zhou YH, Ye M, et al. Burmese injecting drug users in Yunnan play a pivotal role in the cross-border transmission of HIV-1 in the China-Myanmar border region. Virulence. 2018;9(1):1195–1204. doi:10.1080/21505594.2018.1496777
30. Zhou YH, Liang YB, Pang W, et al. Diverse forms of HIV-1 among Burmese long-distance truck drivers imply their contribution to HIV-1 cross-border transmission. BMC Infect Dis. 2014;14(1):463. doi:10.1186/1471-2334-14-463
31. Wan Z, Chen Q, Chen X, et al. HCV diversity among Chinese and Burmese IDUs in Dehong, Yunnan, China. PLoS One. 2016;11(9):e0163062. doi:10.1371/journal.pone.0163062
32. Xuan Q, Liang S, Qin W, et al. High prevalence of HIV-1 transmitted drug resistance among therapy-naive Burmese entering travelers at Dehong ports in Yunnan, China. BMC Infect Dis. 2018;18(1):211. doi:10.1186/s12879-018-3130-9
33. Chen X, Ye M, Duo L, et al. First description of two new HIV-1 recombinant forms CRF82_cpx and CRF83_cpx among drug users in Northern Myanmar. Virulence. 2017;8(5):497–503. doi:10.1080/21505594.2016.1226722
34. Chen X, Ye M, Pang W, Smith DM, Zhang C, Zheng YT. First appearance of HIV-1 CRF07_BC and CRF08_BC outside China. AIDS Res Hum Retroviruses. 2017;33(1):74–76. doi:10.1089/aid.2016.0169
35. Kaye M, Chibo D, Birch C. Phylogenetic investigation of transmission pathways of drug-resistant HIV-1 utilizing pol sequences derived from resistance genotyping. J Acquir Immune Defic Syndr. 2008;49(1):9–16. doi:10.1097/QAI.0b013e318180c8af
36. Chen M, Ma Y, Chen H, et al. HIV-1 genetic transmission networks among men who have sex with men in Kunming, China. PLoS One. 2018;13(4):e0196548. doi:10.1371/journal.pone.0196548
37. Chen M, Ma Y, Duan S, et al. Genetic diversity and drug resistance among newly diagnosed and antiretroviral treatment-naive HIV-infected individuals in western Yunnan: a hot area of viral recombination in China. BMC Infect Dis. 2012;12(1):382. doi:10.1186/1471-2334-12-382
38. Wei H, Xing H, Hsi JH, et al. The sexually driven epidemic in youths in China’s southwestern border region was caused by dynamic emerging multiple recombinant HIV-1 strains. Sci Rep. 2015;5(1):11323. doi:10.1038/srep11323
39. United States Agency for International Development [homepage on the Internet]. Guidelines for the clinical management of HIV infection in Myanmar; 2017. Available from: https://aidsfree.usaid.gov/sites/default/files/mmr_hiv_guidelines_2017.pdf.
40. World Health Organization [homepage on the Internet]. HIV drug resistance report 2017; 2017. Available from: https://www.who.int/hiv/pub/drugresistance/hivdr-report-2017/en/.
41. Yao ST, Chen M, Li YL, et al. HIV drug resistance analysis among Burmese with antiretroviral treatment in Dehong Prefecture, Yunnan Province. Zhonghua Yu Fang Yi Xue Za Zhi. 2016;50(11):1008–1010. doi:10.3760/cma.j.issn.0253-9624.2016.11.016
42. Sanguansittianant S, Nooroon N, Phaengchomduan P, Ammaranond P. Trends in prevalence of HIV-1 drug resistance in Thailand 2009–2010. J Clin Lab Anal. 2013;27(5):346–353. doi:10.1002/jcla.21609
43. Sharma AL, Singh TR, Singh LS. Antiretroviral resistance, genotypic characterization and origin of human immunodeficiency virus among the infected wives of intravenous drug users in Manipur. Sci Rep. 2018;8(1):15183. doi:10.1038/s41598-018-33636-z
44. Liu J, Jia Y, Xu Q, Zheng YT, Zhang C. Phylodynamics of HIV-1 unique recombinant forms in China-Myanmar border: implication for HIV-1 transmission to Myanmar from Dehong, China. Infect Genet Evol. 2012;12(8):1944–1948. doi:10.1016/j.meegid.2012.08.001
45. Chen X, Duo L, Ye M, Zhang C, Zheng YT. Non-Chinese immigrants: challenge faced by Yunnan of China to achieve the 90–90–90 goals. Virol Sin. 2018;33(3):291–293. doi:10.1007/s12250-018-0038-x
46. Liu Y, Li H, Wang X, et al. Natural presence of V179E and rising prevalence of E138G in HIV-1 reverse transcriptase in CRF55_01B viruses. Infect Genet Evol. 2019;77:104098. doi:10.1016/j.meegid.2019.104098
47. Sluis-Cremer N, Jordan MR, Huber K, et al. E138A in HIV-1 reverse transcriptase is more common in subtype C than B: implications for rilpivirine use in resource-limited settings. Antiviral Res. 2014;107:31–34. doi:10.1016/j.antiviral.2014.04.001
48. Smit E, White E, Clark D, et al. An association between K65R and HIV-1 subtype C viruses in patients treated with multiple NRTIs. J Antimicrob Chemother. 2017;72(7):2075–2082. doi:10.1093/jac/dkx091
49. Mackie NE, Dunn DT, Dolling D, et al. The impact of HIV-1 reverse transcriptase polymorphisms on responses to first-line nonnucleoside reverse transcriptase inhibitor-based therapy in HIV-1-infected adults. AIDS. 2013;27(14):2245–2253. doi:10.1097/QAD.0b013e3283636179
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.