Back to Journals » OncoTargets and Therapy » Volume 9
History and progression of Fat cadherins in health and disease
Authors Zhang X, Liu J , Liang X, Chen J, Hong J, Li L, He Q, Cai X
Received 22 April 2016
Accepted for publication 11 October 2016
Published 1 December 2016 Volume 2016:9 Pages 7337—7343
DOI https://doi.org/10.2147/OTT.S111176
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Faris Farassati
Xiaofeng Zhang,1,2,* Jinghua Liu,3,* Xiao Liang,1,2 Jiang Chen,1,2 Junjie Hong,1,2 Libo Li,1 Qiang He,3 Xiujun Cai1,2
1Department of General Surgery, 2Key Laboratory of Surgery of Zhejiang Province, Sir Run Run Shaw Hospital, Zhejiang University, Hangzhou, Zhejiang, 3Department of Hepatobiliary Surgery, Linyi People’s Hospital, Linyi, Shandong, People’s Republic of China
*These authors contribute equally to this work
Abstract: Intercellular adhesions are vital hubs for signaling pathways during multicellular development and animal morphogenesis. In eukaryotes, under aberrant intracellular conditions, cadherins are abnormally regulated, which can result in cellular pathologies such as carcinoma, kidney disease, and autoimmune diseases. As a member of the Ca2+-dependent adhesion superfamily, Fat proteins were first described in the 1920s as an inheritable lethal mutant phenotype in Drosophila, consisting of four member proteins, FAT1, FAT2, FAT3, and FAT4, all of which are highly conserved in structure. Functionally, FAT1 was found to regulate cell migration and growth control through specific protein–protein interactions of its cytoplasmic tail. FAT2 and FAT3 are relatively less studied and are thought to participate in the development of human cancer through a pathway similar to that of the Ena/VASP proteins. In contrast, FAT4 has been widely studied in the context of biological functions and tumor mechanisms and has been shown to regulate the planar cell polarity pathway, the Hippo signaling pathway, the canonical Wnt signaling cascade, and the expression of YAP1. Overall, Fat cadherins may be useful as emerging disease biomarkers and as novel therapeutic targets.
Keywords: Fat cadherins, FAT1, FAT2, FAT3, FAT4, CpG island, Hippo pathway, WNT signaling
Introduction
Cell–cell adhesions are important for the development of multicellular organisms and the process of animal morphogenesis. In eukaryotes, aberrant intracellular environments can induce pathologic regulation of cadherins, leading to cellular pathologies such as carcinoma, kidney disease, and autoimmune diseases.1
As a member of the Ca2+-dependent adhesion superfamily, the cadherin family is divided into several subfamilies: classical cadherins, desmosomal cadherins, protocadherins, Flamingo/Celsr, Dachsous (DS), and Fat cadherins. Each cadherin subfamily exhibits slightly divergent functions. For instance, classical cadherins function as links to the actin cytoskeleton, desmosomal cadherins are specific to the desmosome, protocadherins are associated with stereocilia and somite epithelialization, and Flamingo/Celsr and DS regulate the planar cell polarity (PCP) pathway.2
Fat was first described in the 1920s in Drosophila as an inheritable lethal mutant phenotype.2 Vertebrate Fat cadherin proteins are highly homologous with Drosophila Fat proteins. Four members comprise this gene family: Fat1, Fat2, Fat3, and Fat4.1,3 Sequence analysis of the four vertebrate Fat cadherins, and in particular Fat4, shows that they are orthologous to Drosophila Fat. The Fat family is also well conserved in structure and function, especially in human beings. In the past two decades, the Fat family was demonstrated to be a tumor suppressor.1 In reality, owing to the extremely large size of mRNA transcripts and encoded proteins, understanding the roles of Fat proteins has been challenging, and thus, the roles remain unknown. While both Fat-Hippo and Fat-PCP signaling cascades have previously been shown to result in malformations, tumorigenesis, and cancer metastasis, additional roles remain to be elucidated.3–6 However, with the accumulation of experimental data, technological advancements, and broader knowledge of disease mechanisms, the Fat cadherin family is considered as an “emerging giant” of the cadherin superfamily.
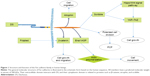
Fat cadherins are single transmembrane receptors, distinguished by 32–34 extracellular cadherin repeats of alternating laminin G-like and EGF-like motifs. FAT1 and FAT4 contain 34 cadherin repeats, while FAT2 and FAT3 contain 32 and 33 repeats, respectively. There are unique arrangements and quantities of EGF-like and laminin G-like motifs that are specific for each member. Usually, the type of protein–protein interaction in the cytoplasmic domain of Fat cadherins differs from that in its functional domains (Figure 1).
Furthermore, because of similarities in the structure of Fat proteins with other characterized proteins, Fat proteins may also act in similar signaling pathways.1–3 In a previous study on the common mutations in Fat genes of patients, nonsynonymous somatic missense mutations did not influence their level of protein expression.7 Additionally, Fat proteins were found to interact with atrophin, a transcriptional co-repressor that also contributes to the function of Fat proteins in the PCP pathway.8
Fat1
Structure and function
Fat1 is located on chromosome 4q34–35 and consists of 27 exons. It is the first Fat family member to be identified and the most frequently studied Fat protein. Fat1 was cloned in 1995 from a human T-leukemia cell line and has since been shown to be closely related to human diseases.1
FAT1 interacts with related cadherins and Ena/VASP proteins to regulate cell migration and actins dynamics. Further studies have shown that FAT1 also binds to Ena/VASP proteins and regulates actin dynamics at both the cell–cell contact and ruffling edges.2,9 Furthermore, a TCF/LEF-binding site was discovered in the Fat1 promoter, 1,100 bp upstream of its transcriptional start site.6
Additionally, because the cytoplasmic region contains various domains, FAT1 can interact directly with the Hippo–Wnt signaling cascade via the atrophin protein, HOMER-1 to HOMER-3 protein, which competes with Ena for binding to FAT1 and β-cadherin.10–12 Finally, FAT1 was shown to undergo alternative splicing in cancer cells undergoing epithelial-to-mesenchymal transition, where its cytoplasmic tail was cleaved by the γ-secretase enzyme complex.13–15
Pathogenicity mechanism and signaling pathways
Analysis of adult human tissues shows Fat1 to be expressed in the development organs and at low levels in adult epithelia.16 Paradoxically, although it is a tumor suppressor, Fat1 was downregulated in invasive breast cancer. On the contrary, Fat1 was upregulated in patients with leukemia and preB-acute lymphoblastic leukemia.17,18 Thus, FAT1 may function as a tumor suppressor and an oncogenic protein.19 Furthermore, FAT1 promotes tumor migration via induction of actin polymerization and shows significant inverse association with the Ki67 index and that loss of membrane localization is correlated with more aggressive tumors.5
However, further studies are required to clarify the role of FAT1. In human diseases, mutations in Fat1 may lead to carcinomas, including cholangiocarcinoma, melanoma, breast cancer, and glioblastoma, typically due to mRNA and protein downregulation or loss of heterozygosity (LOH). In contrast, Fat1 mRNA expression is also upregulated in patients with preB-acute lymphoblastic leukemia (ALL), resulting in shorter lifespan for the patients.20
Furthermore, it is worthwhile to study FAT1 in hepatocellular carcinoma (HCC). Fat1 expression is associated with hypoxia and VEGF in HCC. The hypoxia-induced Fat1 expression may be mediated through expression of MAT2A and subsequent depletion of S-adenosyl-L-methionine levels, resulting in the demethylation of the Fat1 promoter. Incubation with the drug sorafenib in HCC cells results in a significant repression of Fat1 compared to the control.21 However, the relevance of Fat1 in the development and progression of carcinoma is not well understood.
Fat2
Human FAT2 was first discovered in 1998 by motif-trap screening of large molecules containing the EGF motif. Fat2 is closely related to Fat1 and was mapped to a region in human chromosome 5q33.1. The gene encodes a 14.5-kbp mRNA transcript with an open reading frame of 4,349 amino acids.12 In human beings, FAT2 is found to localize at the cell–cell boundaries or periphery of keratinocytes. Furthermore, the disruption of the actin cytoskeleton can perturb cellular FAT2 distribution, as well as FAT1 localization and overlapping dynamic actin networks.
When FAT2 is perturbed, protein domain motifs unique to FAT2 on its cytoplasmic tail become hidden and are unable to bind to EVH1 domains of other proteins. However, an indirect role of FAT2 in regulating actins may possibly depend on interactions with atypical EVH1.13,14 Downregulation of Fat2 via siRNA knockdown in squamous cell carcinoma cells diminished the migration and number of filopodia.15 A Basic Local Alignment Search Tool search of the human Fat2 coding sequence suggests that Fat2 mRNA is expressed in gastric cancer, pancreatic cancer, ovarian cancer, and esophageal cancer. Furthermore, Fat2 expression was detected in hematological tissue, especially in patients with 5q-syndrome. Mutations in Fat2 can also result in the development of head and neck squamous cell cancer.3,6 Whether FAT2 is truly functionally redundant or it significantly changes the pathophysiology in cancer of other organs remains to be elucidated.
Fat3
The third Fat cadherin gene in human, Fat3, found on chromosome 11q14.3-q21 contains 26 exons and encodes a protein of 4,557 amino acids. Further analysis revealed that the Fat3 mRNA pattern of expression was similar to that of Fat1.1,16 The protein FAT3 is analogous to FAT1 in function in relation to interactions with the actin cytoskeleton.
There is evidence suggesting that an EHV1 domain is present in the human FAT3 protein, which may interact with Ena/VASP proteins. However, further studies are required to confirm this.22 Through comparative genomics analysis of the Fat family genes, not only Fat3 was found to be linked with MTNR1B but also the Fat1-MTNR1A and Fat3-MTNR1B loci were paralogous within the human genome. Similarly, comparative proteomics analysis uncovered a C-terminal PDZ-binding motif that was highly conserved among the FAT1 and FAT3 orthologs. However, FAT4 was divergent from other members when studied via phylogenetic analysis.6
In human cancer, a point mutation in Fat3 results in pancreatic tumors. The human surfactant protein C promoter helps to regulate the expression of Fat3, and its expression is downregulated in non-small cell lung cancer. On the contrary, β-catenin and downstream genes of the Wnt signaling pathway are upregulated.23 Whether Fat3 is connected with the Wnt signaling pathway and results in a better prognosis of carcinoma remain controversial.
Fat4
Structure and interacting proteins
The Fat4 cadherin gene is located on chromosome 4q28.1 and has 17 known exons. When transcribed, the mRNA is ~16 kb and encodes a protein of 4,924 amino acids. The cytoplasmic structure of FAT4 is highly conserved as compared to the other Fat family proteins. Owing to sequence similarities and correlation with known functional pathways, the role and function of FAT3 may be homologous with those of other species.1 Furthermore, the ubiquitous expression of Fat proteins in major tissues appears to be divided into a pattern of FAT1/FAT3 and FAT4 in the epithelial and mesenchymal tissues, respectively. This evidence suggests that the function of FAT4 is conserved among mammals and thus supports a role in the PCP pathway. The protein DCSH1 functions as a ligand for FAT4 and may be regarded as a reciprocal Fat-to-DCSH1 signal.5,19 There is evidence suggesting FAT4 to be strongly PARsylated by TNKS. This is because FAT4 was discovered to contain a TNKS-binding motif using an anti-poly(ADP-ribose) antibody to label PARsylated proteins. Other TNKS targets, such as DISC1, warrant further investigation owing to structural and sequence similarities to FAT4. Interestingly, the endogenous expression of DISC1 and FAT4 is undetectable in HEK293T cells.18
Signaling pathways and pathogenicity mechanism
The core components of the Hippo signaling pathway are SAV, Wnts, Hippo, and YKI and are involved in controlling organ size.20–22 Fat proteins may function at the apical point of the pathway as a single transmembrane receptor. FAT4 belongs to the E-cadherin family and mediates Wnt and PCP signaling cascades through the non-canonical Wnt signaling pathway, which is considered to play crucial roles in tissue homeostasis, vascularization, tumorigenesis, invasion, and metastasis.24–26 However, E-cadherin has not been shown to be associated with tumor invasion, cancer, or metastasis.27–29 The known roles of FAT4 include inhibiting proliferation, promoting colony formation, and reducing fibronectin adhesion.
Interestingly, there is an increasing body of evidence suggesting conserved DS/Fat-PCP signaling mechanisms, due to the ability of human FAT4 to rescue PCP mutant phenotypes and aberrant cellular polarization.5 Furthermore, by quantifying the evolution of the cancer genome using the gene gravity model in several patients with cancer, six putative genes were identified, and among this list is Fat4. Four of these identified genes (Fat4, SYNE1 AHNAK, and COLL11A1) not only contained missense mutations but also showed a significantly higher mutation density in the cancer genome at the whole-genome level.30
On the other hand, the Fat4 gene was methylated in 39% of tumors, similar to the components of the Hippo growth control pathway. Furthermore, hypermethylation of CpG islands represents malignant progression. Thus, the methylation of Fat4 may be regarded as a potential epigenetic “driver” event for the development of tumors, rather than just a “passenger” event that would be without functional sequences.31 Moreover, Fat4 was silenced by methylation at CpG islands and was associated with infection by Helicobacter pylori in gastric cancer. Surprisingly, only the CpG island methylator phenotype was associated with clinicopathological characteristics.27
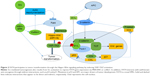
In addition, FAT4 mutations are usually located in the EGF-like domain, transmembrane domain (p.Pro4454Leu), cadherin repeat domain (p.Leu1062Arg), and even the cytoplasmic region (p.Gln4872His). In one instance where FAT4 contained a p.Asp4101Tyr mutation, the protein showed adverse interactions, and its function of inhibiting malignancies was compromised (Figure 3).24,32 The FAT4 mutations, which inhibit cell differentiation, were attributed to the activation of the Hippo pathway through the transcriptional co-activator proteins YAP and TAZ; however, the molecular pathway connecting FAT4 to YAP does not appear to be conserved in mammals.5,33
Data from genome-wide association studies and exome sequencing identified Fat4 as a candidate gene for tumorigenic events in lung adenomas and gastric cancer.30,33,34 Recent evidence has also shown Fat4 mRNA to be downregulated by Src through the MEK–ERK pathway, where Src-mediated actin depolymerization has been hypothesized to downregulate Fat4 expression via hypermethylation of the DNMT-1 promoter.34,35 Surprisingly, Fat4 mRNA expression was also shown to be associated with mechanical stress that stems from the extracellular environment.36,37 In addition, FAT4 regulates actin structure and represses connective tissue growth factor (CTGF) mRNA expression through inhibition or inactivation of the YAP–TAZ pathway.38,39 A recent study showed that when EBLN1 is silenced, the downregulation of Fat4 may influence the cell cycle, ubiquitin hydrolysis, mitogen-activated protein kinase, p53, and apoptosis (Figure 2).40 Further studies are required to elucidate the pathway or mechanisms involved.
Correlation of human tumor
In human tumors, Fat4 is recurrently mutated in several types of human cancers such as melanoma, gastric cancer, pancreatic cancer, colorectal cancer, and HCC. Fat4 exhibited a mutation rate of >10% and was classified as a novel somatic mutation gene in colorectal cancer and chronic myelomonocytic leukemia.41,42 Likewise, promoter hypermethylation or LOH of Fat4 has been reported in breast and lung cancers.43,44 Epigenetic and genetic mechanisms are involved in the disruption of FAT4 function, and knockdown of Fat4 can reduce cell adhesion, as well as strongly induce cell migration and invasion in gastric cancer.2,3,8
A screen to study the prevalence of mutations in Fat4 showed that ~5% of mutations were in CpG islands, 4% of which were genomic deletions.22 Surprisingly, the protein expression levels of FAT4 have not been examined in gastric adenocarcinoma (Figure 3).10 However, tumor growth is inhibited by reintroduction of a wild-type Fat4 DNA sequence into cells derived from cutaneous tumor.3 Strangely, FAT4 may also have a significant influence on the formation of pulmonary adenoma in males compared to that in females.45 In patients with pre-B ALL, FAT4 directly interacts with MPDZ and MUPP1 to recruit membrane-associated guanylate kinase MPP5/PALS1.18,46,47 The KSHV gene is one of the direct downstream targets of miR31 and is also involved in the reduction of FAT4-enhanced tumor cell motility.48 In chronic diseases, such as coronary artery disease, FAT4 inhibits miR720 expression via repressing PCP genes. Furthermore, miR31 regulates endothelial progenitor cell function and expression of miR720 through the suppression of Fat4 and related planer polarity signaling genes, respectively.49
Conclusion
Despite the fact that the Fat family was discovered and studied nearly a century ago, these “giant genes” and proteins have often been overlooked due to their large size. However, with the development of next-generation sequencing, studies have revealed intriguing biological functions for the Fat proteins. In particular, ectopic Fat cadherin expression is intimately linked to human diseases.1 Among them is FAT1 interacting with Ena/VASP proteins to regulate actin dynamics at both the cell–cell contact and leading edges.2 Studies have shown that FAT1 is involved in the regulation of cell migration and the growth control through specific protein–protein interactions of its cytoplasmic tail.1 FAT2 and FAT3 are relatively less studied and presumed to participate in the same pathway as Ena/VASP in the development of human cancers such as pancreatic cancer and gastric cancer.3,6 The FAT4 protein is different from others since it has been widely studied in biological function and tumor mechanisms to regulate the PCP pathway, the Hippo signaling pathway, the canonical Wnt signaling cascade, and expression of YAP1.3,8,25,26
Currently, it is unknown whether any of the genetic changes observed in the Fat family can significantly affect the pathophysiology of cancer due to epigenetic changes in CpG islands and genetic mutations during multi-stage carcinogenesis.1,6 Additionally, FAT4 is conserved between arthropods and vertebrates. As a tumor suppressor gene, research on FAT4 may be applied toward diagnostics, prognostics, and therapeutics in the era of personalized medicine. From an evolutionary point of view, any genetic change suggests that the progress is revolutionary, such that the Fat cadherins may be regarded as emerging disease biomarkers and as targets for novel therapies or surveillance.
Acknowledgments
This study was supported by the International Scientific and Technological Cooperation Projects (2012DFA30410), the Science-technology Projects of Zhejiang Province (LY15H160043), and the National Science-technology Support Plan Projects (2012BAI14B06). We thank Wenbin Jiang, Jiacheng Tang, and Xu Feng for revising the figures and writing and advising, in the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Sadeqzadeh E, de Bock CE, Thorne RF. Sleeping giants: emerging roles for the fat cadherins in health and disease. Med Res Rev. 2014;34(1):190–221. | ||
Tanoue T, Takeichi M. New insights into fat cadherins. J Cell Sci. 2005;118(pt 11):2347–2353. | ||
Katoh M. Function and cancer genomics of fat family genes. Int J Oncol. 2012;41(6):1913–1918. | ||
Alders M, Al-Gazali L, Cordeiro I, et al. Hennekam syndrome can be caused by FAT4 mutations and be allelic to Van Maldergem syndrome. Hum Genet. 2014;133(9):1161–1167. | ||
Mao Y, Francis-West P, Irvine KD. Fat4/Dchs1 signaling between stromal and cap mesenchyme cells influences nephrogenesis and ureteric bud branching. Development. 2015;142(15):475–481. | ||
Katoh Y, Katoh M. Comparative integromics on FAT1, FAT2, FAT3 and FAT4. Int J Mol Med. 2006;18(3):523–528. | ||
Furukawa T, Sakamoto H, Takeuchi S, et al. Whole exome sequencing reveals recurrent mutations in BRCA2 and Fat genes in acinar cell carcinomas of the pancreas. Sci Rep. 2015;5:8829. | ||
Qi C, Zhu YT, Hu L, Zhu YJ. Identification of Fat4 as a candidate tumor suppressor gene in breast cancers. Int J Cancer. 2009;124(4):793–798. | ||
Badouel C, Zander MA, Liscio N, et al. Fat1 interacts with Fat4 to regulate neural tube closure, neural progenitor proliferation and apical constriction during mouse brain development. Development. 2015;142(16):535–540. | ||
Jung HY, Cho H, Oh MH, et al. Loss of fat atypical cadherin 4 expression is associated with high pathologic T stage in radically resected gastric cancer. J Gastric Cancer. 2015;15(1):39–45. | ||
Sadeqzadeh E, de Bock CE, Zhang XD, et al. Dual processing of Fat1 cadherin protein by human melanoma cells generates distinct protein products. J Biol Chem. 2011;286(32):28181–28191. | ||
Nakayama M, Nakajima D, Nagase T, Nomura N, Seki N, Ohara O. Identification of high-molecular-weight proteins with multiple EGF-like motifs by motif-trap screening. Genomics. 1998;51(1):27–34. | ||
Moeller MJ, Soofi A, Braun GS, et al. Protocadherin Fat1binds Ena/VASP proteins and is necessary for actin dynamics and cell polarization. EMBO J. 2004;23(19):3769–3779. | ||
Tanoue T, Takeichi M. Mammalian Fat1 cadherin regulates actin dynamics and cell-cell contact. J Cell Biol. 2004;165(4):517–528. | ||
Matsui S, Utani A, Takahashi K, et al. Knockdown of Fat2 by siRNA inhibits the migration of human squamous carcinoma cells. J Dermatol Sci. 2008;51(3):207–210. | ||
Mitsui K, Nakajima D, Ohara O, Nakayama M. Mammalian Fat3: a large protein that contains multiple cadherin and EGF-like motifs. Biochem Biophys Res Commun. 2002;290(4):1260–1266. | ||
Kwaepila N, Burns G, Leong AS. Immunohistological localisation of human FAT1(hFAT) protein in 326 breast cancers. Does this adhesion molecule have a role in pathogenesis? Pathology. 2006;38(2):125–131. | ||
de Bock CE, Ardjmand A, Molloy TJ, et al. The Fat1 cadherin is overexpressed and an independent prognostic factor for survival in paired diagnosis-relapse samples of precursor B-cell acute lymphoblastic leukemia. Leukemia. 2012;26(5):918–926. | ||
Rock R, Schrauth S, Gessler M. Expression of mouse dchs1, fjx1, and Fat-j suggests conservation of the planar cell polarity pathway identified in Drosophila. Dev Dyn. 2005;234(3):747–755. | ||
Guettler S, LaRose J, Petsalaki E, et al. Structural basis and sequence rules for substrate recognition by Tankyrase explain the basis for cherubism disease. Cell. 2011;147(6):1340–1354. | ||
Rauch TA, Wang Z, Wu X, Kernstine KH, Riggs AD, Pfeifer GP. DNA methylation biomarkers for lung cancer. Tumour Biol. 2012;33(2):287–296. | ||
Deans MR, Krol A, Abraira VE, Copley CO, Tucker AF, Goodrich LV. Control of neuronal morphology by the atypical cadherin Fat3. Neuron. 2011;71(5):820–832. | ||
Rohrbeck A, Borlak J. Cancer genomics identifies regulatory gene networks associated with the transition from dysplasia to advanced lung adenocarcinomas induced by c-Raf-1. PLoS One. 2009;4(10):e7315. | ||
Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21(8):886–897. | ||
Tapon N, Harvey KF, Bell DW, et al. Salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110(4):467–478. | ||
Lin Y, Wu Z, Guo W, Li J. Gene mutations in gastric cancer: a review of recent next-generation sequencing studies. Tumour Biol. 2015;36(10):7385–7394. | ||
Yoshida S, Yamashita S, Niwa T, et al. Epigenetic inactivation of FAT4, contributes to gastric field cancerization. Gastric Cancer. Epub 2016 Jan 20. | ||
Czyzewska J, Guzińska-Ustymowicz K, Ustymowicz M, Pryczynicz A, Kemona A. The expression of E-cadherin-catenin complex in patients with advanced gastric cancer: role in formation of metastasis. Folia Histochem Cytobiol. 2010;48(1):37–45. | ||
Chen HC, Chu RY, Hsu PN, et al. Loss of E-cadherin expression correlates with poor differentiation and invasion into adjacent organs in gastric adenocarcinomas. Cancer Lett. 2003;201(1):97–106. | ||
Cheng F, Liu C, Lin CC, et al. A gene gravity model for the evolution of cancer genomes: a study of 3,000 cancer genomes across 9 cancer types. PLoS Comput Biol. 2015;11(9):e1004497. | ||
Valletta D, Czech B, Spruss T, et al. Regulation and function of the atypical cadherin FAT1 in hepatocellular carcinoma. Carcinogenesis. 2014;35(6):1407–1415. | ||
Tenedini E, Bernardis I, Artusi V, et al. Targeted cancer exome sequencing reveals recurrent mutations in myeloproliferative neoplasms. Leukemia. 2014;28(5):1052–1059. | ||
Bossuyt W, Chen CL, Chen Q, et al. An evolutionary shift in the regulation of the Hippo pathway between mice and flies. Oncogene. 2014;33(10):1218–1228. | ||
Zang ZJ, Cutcutache I, Poon SL, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44(5):570–574. | ||
Wang K, Kan J, Yuen ST, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43(12):1219–1223. | ||
Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. | ||
Nam JS, Ino Y, Sakamoto M, Hirohashi S. Src family kinase inhibitor PP2 restores the E-cadherin/catenin cell adhesion system in human cancer cells and reduces cancer metastasis. Clin Cancer Res. 2002;8(7):2430–2436. | ||
Dupont S, Morsut L, Aragona M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. | ||
Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11(5):353–365. | ||
Peng H, Lin S, Dan Z, et al. Knock-down of endogenous bornavirus-like nucleoprotein 1 inhibits cell growth and induces apoptosis in human oligodendroglia cells. Int J Mol Sci. 2016;17(4):435. | ||
Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19(4):491–505. | ||
Mason CC, Khorashad JS, Tantravahi SK, et al. Age-related mutations and chronic myelomonocytic leukemia. Leukemia. 2016;30(4):906–913. | ||
Badouel C, McNeill H. SnapShot: the hippo signaling pathway. Cell. 2011;145:484–484. | ||
Yu J, Wu WK, Li X, et al. Novel recurrently mutated genes and a prognostic mutation signature in colorectal cancer. Gut. 2015;64(4):636–645. | ||
Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299(3):551–572. | ||
Yamamoto H, Watanabe Y, Maehata T, et al. An updated review of gastric cancer in the next-generation sequencing era: insights from bench to bedside and vice versa. World J Gastroenterol. 2014;20(14):3927–3937. | ||
Berndt A, Cario CL, Silva KA, et al. Identification of Fat4 and tsc22d1 as novel candidate genes for spontaneous pulmonary adenomas. Cancer Res. 2011;71(17):5779–5791. | ||
Wu YH, Hu TF, Chen YC, et al. The manipulation of miRNA-gene regulatory networks by KSHV induces endothelial cell motility. Blood. 2011;118(10):2896–2905. | ||
Wang HW, Huang TS, Lo HH, et al. Deficiency of the microRNA-31-microRNA-720 pathway in the plasma and endothelial progenitor cells from patients with coronary artery disease. Arterioscler Thromb Vasc Biol. 2014;34(4):857–869. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.