Back to Journals » OncoTargets and Therapy » Volume 10
Histone demethylase KDM2B upregulates histone methyltransferase EZH2 expression and contributes to the progression of ovarian cancer in vitro and in vivo
Authors Kuang Y, Lu F, Guo J, Xu H, Wang Q, Xu C, Zeng L, Yi S
Received 15 February 2017
Accepted for publication 14 April 2017
Published 26 June 2017 Volume 2017:10 Pages 3131—3144
DOI https://doi.org/10.2147/OTT.S134784
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Carlos E Vigil
Yan Kuang,1 Fangfang Lu,1 Jianfeng Guo,2 Hong Xu,1 Qi Wang,1 Chaohuan Xu,1 Longjia Zeng,1 Suyi Yi1
1Department of Obstetrics and Gynecology, First Affiliated Hospital of Guangxi Medical University, Nanning, 2Department of Obstetrics and Gynecology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, People’s Republic of China
Abstract: Aberrant histone methylation contributes to the progression and development of many tumors. Histone methylation is a dynamic process regulated by both histone demethylase and histone methyltransferase, which ultimately alters the levels of gene transcription. However, the relationship between histone demethylase and histone methyltransferase, as well as their regulatory mechanisms in ovarian cancer development, is still unclear. Lysine-specific demethylase 2B (KDM2B) is a key demethylase of H3K36me3 and H3K4me3 that regulates gene expression and plays a role in tumorigenesis via epigenetic mechanisms. To determine the expression pattern of KDM2B in ovarian neoplasms, we analyzed the mRNA and protein levels of KDM2B and the histone methyltransferase enhancer of zester homolog 2 (EZH2) in normal, benign, borderline, and malignant ovarian tissue samples. We found that KDM2B expression was gradually increased in ovarian tumors, with the highest expression found in the malignant ovarian tissues, and the differences in KDM2B expression among the different International Federation of Gynecology and Obstetrics stages and pathological grades/types were statistically significant. Moreover, KDM2B expression was positively correlated with EZH2 expression in ovarian tissues. To determine the role of KDM2B in tumorigenesis in vitro and in vivo, we silenced KDM2B expression in ovarian cancer cells using the KDM2B short hairpin RNA expression lentivirus and established a nude mouse xenograft model. Downregulation of endogenous KDM2B decreased the expression of EZH2 and reduced the proliferation and migration of ovarian cancer cells. Loss of KDM2B suppressed ovarian tumor formation in vivo. Our results suggest that KDM2B plays an important role in the tumorigenesis of ovarian cancer, with a possible mechanism of increasing the expression of the oncogene EZH2; this indicates that certain histone methyltransferase may be positively regulated by certain histone demethylase in the epigenetic regulation of ovarian tumors. KDM2B may be a novel therapeutic target for the clinical treatment of ovarian cancer.
Keywords: ovarian cancer, KDM2B, EZH2, epigenetic regulation, tumor formation
Introduction
Epigenetic mechanisms, including DNA methylation, non-coding RNA regulation, chromatin remodeling, and histone modification, have been demonstrated to be involved in regulating many normal physiological functions of eukaryotic cells. Therefore, epigenetic changes in any part of cellular regulation may lead to cell functional abnormality and cause many diseases.1 In some studies, tumor suppressor gene hypermethylation, genome-wide loss of DNA methylation, and histone modification were considered to be closely related to tumorigenesis.2,3 Studies have reported that aberrant histone modification plays important roles in the progression and development of many tumors, which have been characterized by abnormal methylation/acetylation of amino acid residues in histones.4,5
Histone methylation is a dynamic process that can influence the interaction of histones with other functional proteins by dynamic regulation between histone demethylase and histone methyltransferase, ultimately altering the level of gene transcription.6 However, the relationship between histone demethylase and histone methyltransferase, as well as their regulatory mechanism in tumor development, is still unclear.
The oncogenic potential epigenetic regulator lysine-specific demethylase 2B (KDM2B), which is a member of the JmjC domain-containing histone demethylase (JHDM) family, has recently drawn a great deal of attention.7,8 KDM2B regulates gene transcription via the demethylation of dimethyl histone H3 lysine 36 (H3K36me2) and trimethyl histone H3 lysine 4 (H3K4me3).9 Some studies have indicated that the abnormal expression of KDM2B inhibits tumor suppressor genes and promotes oncogene expression, thereby contributing to uncontrolled cell growth and possibly leading to tumorigenesis.10,11 However, the roles of histone demethylase KDM2B during ovarian cancer progression are still unclear.
Enhancer of zester homolog 2 (EZH2) is a key enzyme subunit of polycomb repressive complex 2 that acts as a histone methyltransferase of histone H3 lysine 27 (H3K27). The methylated H3K27 leads to epigenetic silencing of target genes.12 Multiple studies have shown that EZH2 is an oncoprotein involved in the progression of many malignant tumors.13–15 In breast and ovarian cancers, highly expressed EZH2 is associated with a poor prognosis for patients.16,17 In our previous study, we found that EZH2 expression was significantly increased in ovarian carcinoma compared with normal tissues. Moreover, EZH2 blocking suppressed the migration and invasion capability of ovarian cancer cells both in vitro and in vivo.18
Karoopongse et al found that EZH2 expression in myelodysplastic syndrome cells was decreased after KDM2B knockdown.19 This result suggests that EZH2 expression may be regulated by KDM2B. Therefore, we hypothesize that the histone demethylase KDM2B can regulate histone methyltransferase EZH2 expression and contribute to the progression of ovarian cancer. To prove our hypothesis, we examined KDM2B and EZH2 expression in normal ovarian tissue, benign tumor, borderline tumor, and ovarian carcinoma and analyzed the correlation of their expression with the clinicopathological characteristics of ovarian carcinoma and the relationship between KDM2B and EZH2 expression. Furthermore, we demonstrated the role of KDM2B in the regulation of EZH2 gene expression, as well as in the proliferation and migration of ovarian cancer cells in vitro and in vivo; the preliminary results revealed potential regulatory relationships between certain histone demethylases and certain histone methyltransferases in ovarian cancer.
Materials and methods
Ovarian tissue samples
A total of 153 ovarian tissue samples were obtained from patients who underwent surgery between December 2009 and June 2015 in First Affiliated Hospital of Guangxi Medical University (Nanning, People’s Republic of China). The samples included 20 cases of normal ovarian tissue, 33 cases of benign ovarian tumor (15 serous cystadenomas and 18 mucous cystadenomas), 21 cases of borderline tumor (14 serous cystadenomas and 7 mucous cystadenomas), 70 cases of epithelial ovarian cancer (42 serous adenocarcinomas, 20 mucous adenocarcinomas, 6 endometrioid carcinomas, and 1 squamous carcinoma), and 10 cases of non-epithelial ovarian cancer (4 granulosa cell tumors, 2 immature teratomas, 1 thecoma, 1 yolk sac tumor, 1 dysgerminoma, and 1 androblastoma).
The normal ovarian specimens were from patients with uterine myoma or adenomyosis who underwent adnexectomy. The histological type of all specimens was confirmed by pathological examination. None of the patients underwent chemotherapy or radiotherapy before the surgery, and the surgical staging of malignant tumor was determined according to the International Federation of Gynecology and Obstetrics (FIGO) guidelines. All the enrolled patients signed written informed consent forms for this study, and the study was approved by the Institutional Research Ethics Committee of Guangxi Medical University.
Cell lines
The ovarian cancer cell lines, SKOV3 and A2780, were obtained from the China Center for Type Culture Collection (CCTCC, Wuhan, People’s Republic of China). A2780 cells were derived from a patient with primary ovarian adenocarcinoma, and SKOV3 cells were derived from malignant ascites of a patient with aggressive ovarian cancer. Both cell lines were cultured in RPMI-1640 media (Hyclone, Logan, UT, USA) with 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA) and maintained at 37°C in a humidified atmosphere of 5% CO2.
Immunohistochemistry
Formalin-fixed, paraffin-embedded tissue sections were used for immunohistochemistry (IHC) analysis. Ovarian tissue sections were deparaffinized with xylene and rehydrated in graded ethanol. Antigen retrieval was performed by boiling tissue sections in 10 mM sodium citrate buffer (pH 6.0) for 5 min. Following this, the sections were immersed in 3% H2O2 for 10 min to inactivate endogenous peroxidase. After non-specific antigen sites had been blocked, tissue sections were incubated with goat anti-KDM2B antibody (1:100 dilutions; Abcam, Cambridge, MA, USA) or rabbit anti-EZH2 antibody (1:50 dilution; Abcam) overnight at 4°C. Immunostaining was performed with diaminobenzidine using the streptavidin–biotin complex/horseradish peroxidase method, and hematoxylin was used for counterstaining.
IHC staining analysis
Positive expression was defined as cells presenting brownish yellow granules located in the nucleus and cytoplasm. Five visual fields containing the most concentrated positive cells were observed under a ×100 optical microscope. One hundred cells in the same fields under a ×400 optical microscope were chosen for calculating the staining intensity and average percentage of positive cells in each field (×400). The staining intensity was scored as follows: non-staining, light yellow, brownish yellow, and dark brown colors were graded as 1, 2, 3, and 4, respectively. For the average percentages of positive cells, 0–25%, 26%–50%, 51%–75%, and more than 75% were scored as 1, 2, 3, and 4, respectively. The scores of staining intensity and the average percentage of positive cells were multiplied, and scores of 0–4, 4–8, 8–12, and 12–16 were considered negative (−), weak (+), moderate (++), and strong (+++), respectively. Weak, moderate, and strong expressions were all considered to represent positive expression.
KDM2B short hairpin RNA lentiviral construction and cell transduction
KDM2B short hairpin RNA (shRNA) was designed and synthesized according to the KDM2B gene sequences (GenBank Accession Number: NM_032590). The sequence of KDM2B siRNA oligonucleotides was 5′-TTCTTCAAACGCTGTGGAA-3′. The sequence of negative control siRNA oligonucleotides was 5′-TTCTCCGAACGTGTCACGT-3′. After annealing reaction in a polymerase chain reaction (PCR) instrument, shKDM2B and shNC fragments were cloned into an shRNA expression vector (Genechem, Shanghai, People’s Republic of China) and used for lentivirus packaging. Lentiviral transduction of A2780 and SKOV3 cells was performed according to the manufacturer’s protocol.
RNA isolation and real-time quantitative PCR
Total RNA was extracted from ovarian tissue samples or cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. KDM2B and EZH2 cDNA were synthesized using a PrimeScript™ RT reagent kit (TAKARA Bio Inc., Kyoto, Japan). The forward and reverse primer sequences for KDM2B mRNA detection were 5′-CTCACTGCTGTTGGCACCAC-3′ and 5′-TGCTTGCAGTACCTCAGGTCAATA-3′, respectively. The forward and reverse primer sequences for EZH2 mRNA detection were 5′-TTGTTGGCGGAAGCGTGTAAAATC-3′ and 5′-TCCCTAGTCCCGCGCAATGAGC-3′, respectively. The sequences of the β-actin forward and reverse primers were 5′-GTCCACCGCAAATGCTTCTA-3′ and 5′-TGCTGTCACCTTCACCGTTC-3′, respectively. The real-time PCR reaction was performed in an Applied Biosystems 7300 Real-time PCR system (Applied Biosystems, Foster City, CA, USA).
The final volume of each reaction was 20 μL, containing 0.6 μL of 10 μM primer, 1.5 μL of cDNA sample, 7.3 μL of RNase-free water, and 10 μL of SYBR-Green Real-time PCR Master Mix (Rox; Roche Diagnostics GmbH, Mannheim, Germany). Reactions were performed according to the following cycling conditions: initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 1 min. The relative KDM2B and EZH2 mRNA levels were calculated using the comparative threshold cycle (Ct) method (2−ΔΔCt) normalized by β-actin expression. All experiments were repeated three times.
Protein extraction and Western blot
Total cell lysates of A2780 and SKOV3 cells were obtained using radioimmunoprecipitation assay buffer (Millipore, Billerica, MA, USA) according to the manufacturer’s protocol. The total protein concentrations of each sample were measured via bicinchoninic acid assay (Beyotime, Shanghai, People’s Republic of China). Forty micrograms of total proteins were denatured in boiling water for 5 min and separated with 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis. The separated proteins were transferred onto polyvinylidene fluoride membrane, blocked with freshly prepared 5% bovine serum albumin for 2 h, and then incubated with rabbit anti-KDM2B polyclonal antibody (1:1,000 dilution; Millipore) or rabbit anti-EZH2 polyclonal antibody (1:1,000 dilution; Abcam) for 24 h at 4°C. Fluorescent dye-conjugated anti-rabbit secondary antibody (1:15,000 dilution; Cell Signaling, Danvers, MA, USA) was used for detecting the primary antibodies for 1 h at room temperature. After washing three times with TBST, protein bands were visualized using the Odyssey infrared fluorescence scanning imaging system (LI-COR Biosciences, Lincoln, NE, USA).
Cell proliferation assay
Cell proliferation assay was performed using a cell counting kit-8 (CCK-8) (Dojindo, Kumamoto, Japan) according to the manufacturer’s instructions. Briefly, 48 h after lentivirus transfection, A2780 and SKOV3 cells were seeded into 96-well plates at a density of 3,000 cells/well in sextuplicate and cultured at 37°C in a 5% CO2 atmosphere. CCK-8 was added into the 96-well plates and incubated for 1 h; the optical densities of each well were then detected at 450 nm in a microplate reader every 24 h for 5 days. Cell growth curves were generated by calculating the average optical density of each day.
Wound healing assay
A wound healing assay was performed to analyze the migration ability of ovarian cancer cells. A2780 or SKOV3 cells (1×104 cells/well) were seeded into 96-well plates and cultured at 37°C in 5% CO2 atmosphere. After a confluent cell layer had formed, a 10 μL tip was used to draw a line at the bottom of the 96-well plates. Cells were washed with phosphate-buffered saline (PBS) and cultured in serum-free medium. Scratch widths in the same visual field were measured at 0, 12, 24, and 48 h. The experiment was performed in triplicate.
Transwell migration assay
For transwell migration assay, cells (1×106/well) were resuspended with 100 μL of serum-free RPMI-1640 medium after trypsinization and then seeded into the upper chambers of the transwell inserts (Corning Costar, Cambridge, MA, USA). Following this, 600 μL of RPMI-1640 complete medium was added to the lower chambers, and the plates were incubated for 24 h at 37°C in a 5% CO2 atmosphere. Migrated cells on the bottom surface of the membrane were fixed with methanol and stained with 0.1% crystal violet. Cell numbers were counted after randomly selecting 5 high-power fields (×400) under the microscope, and the average number of cells was used for assessing migration ability. All assays were repeated three times.
Cell cycle analysis
A2780 cells were trypsinized, collected by centrifugation at 1,000 g for 5 min, and fixed with 70% ethanol at 4°C for 3 h after washing twice with PBS. The fixed cells were washed with PBS, resuspended in 100 μL of RNase A, and incubated for 30 min at 37°C. After the cells had been incubated with 400 μL of propidium iodide in the dark for 30 min at 4°C, the cell cycle was analyzed using flow cytometry.
In vivo cancer growth of ovarian cancer in a xenograft model
An ovarian carcinoma xenograft model was employed to investigate the effect of KDM2B downregulation in tumorigenesis. Ten 5-week-old female nude mice were divided into two groups (five mice/group), which were subcutaneously injected with 1×106 shKDM2B-A2780 cells or shNC-A2780 cells into the dorsal flank, respectively. Tumor volumes were measured every week, and after the mice had developed cachexia symptoms, xenografts were removed for further study. Each tumor was analyzed for KDM2B and EZH2 mRNA and protein levels by real-time quantitative PCR (RT-qPCR) and Western blot analysis. This animal experiment was approved by the Animal Experimental Ethics Committee of Guangxi Medical University. The care of laboratory animals and animal experimentation have conformed to the administration rules of laboratory animals of the Animal Experimental Ethics Committee of Guangxi Medical University.
Statistical analyses
All data were processed using SPSS 13.0 statistic software (IBM Corp., Armonk, NY, USA). The data were evaluated using analysis of variance and Student’s t-test. The chi-square test and Fisher’s exact test were employed to assess the association between KDM2B and EZH2 expression and clinicopathological characteristics (age, histological type, tumor grade, FIGO stage, and lymph node metastasis) of ovarian cancer. Correlations between KDM2B and EZH2 mRNA expression were assessed using Pearson’s correlation test. P<0.05 was defined as statistically significant.
Results
KDM2B and EZH2 were highly expressed in ovarian cancer
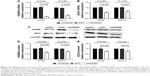
To determine the expressions of KDM2B and EZH2 in ovarian carcinoma, IHC was performed on 153 ovarian tissue specimens. Figure 1A shows the representative IHC staining of KDM2B and EZH2 in normal, benign, borderline, and malignant tissue specimens. KDM2B expression was observed in 10.00% of normal ovarian tissue specimens (2/20), 50.00% of benign tumors (16/32), 63.64% of borderline tumors (14/22), and 73.75% ovarian cancer (59/80), with a statistical significance of P<0.001 (χ2=28.332). Moreover, EZH2 expression was positive in 72.50% (58/80) ovarian cancer, which was higher than the observed 63.64% (14/22) in borderline tumors, the 56.25% (18/32) in benign tumors, and the 20% (4/20) in normal tissues. There was a statistically significant difference among the groups (χ2=18.954, P<0.001; Table 1).
  | Table 1 KDM2B and EZH2 protein expression in different ovarian tissue specimens |
Fluorescent RT-qPCR was used for determining mRNA levels of KDM2B and EZH2 in 12 normal ovarian tissue specimens, 15 benign tumors, 10 borderline neoplasms, and 25 ovarian carcinomas, which were randomly selected. We observed that compared with normal ovarian tissue samples, KDM2B mRNA expression level in benign, borderline, and malignant tissue samples was increased by 2-fold (21.19), 12-fold (23.62), and 20-fold (24.32), respectively (P<0.001). Similarly, EZH2 mRNA expression level was increased by 1.6-fold (20.66) in benign tumors, 2.4-fold (21.25) in borderline tumors, and 3.9-fold (21.95) in malignant tumors (P<0.001). There were significant differences in KDM2B and EZH2 expression levels among normal ovaries, benign tumors, borderline tumors, and ovarian cancers (Figure 1B and C).
EZH2 expression was positively correlated to KDM2B
The correlation between KDM2B and EZH2 was further analyzed using Pearson correlation analysis. We observed that high expression of KDM2B contributed to the increased level of EZH2, and EZH2 was positively associated with KDM2B in 62 randomly selected ovarian samples, with a statistical significance level of P<0.001 (r=0.832; Figure 1D).
KDM2B and EZH2 expression is associated with the clinicopathological features of ovarian carcinoma patients
We analyzed the relationship between KDM2B or EZH2 expression and the clinicopathological features of malignant tissues from 80 ovarian carcinoma patients (Table 2). Our results indicated that KDM2B was expressed in 78.57% (55/70) of epithelial ovarian and 40% (4/10) of non-epithelial ovarian cancers (P=0.018). The positive expression of KDM2B was found in 58.54% (24/41) of the moderately and highly differentiated (G1/G2) ovarian cancer patients and in 89.74% (35/39) of the poorly differentiated (G3) ovarian cancer patients (χ2=10.055, P=0.002). Meanwhile, KDM2B expression was found in 92.11% (35/38) of patients with stages III/IV, representing a higher proportion than that found in patients with stages I/II, where it was expressed in 57.14% (24/42) of patients (χ2=12.597, P<0.001). In terms of lymph node metastasis, KDM2B expression was detected more frequently in the group with lymph node metastasis (90.63%, 29/32) than in the group without lymph node metastasis (62.50%, 30/48; χ2=7.845, P=0.005).
Similarly, the EZH2-positive rates in epithelial and non-epithelial ovarian carcinoma were 77.14% (54/70) and 40% (4/10), respectively (P=0.023). In G3-differentiated cancer, the EZH2-positive rate was 92.31% (36/39), which was higher than in G1/G2-differentiated cancer, where the EZH2-positive rate was 56.10% (23/41). The difference was statistically significant (χ2=13.537, P<0.001). The expression of EZH2 was observed in 50% (21/42) of patients in stages I/II, while in the patients with III/VI stages, the EZH2-positive rate was 97.37% (37/38; χ2=22.452, P<0.001). For lymph node metastasis, the EZH2-positive rate in the patients without metastasis was 58.33% (28/48), and this increased to 93.75% (30/32) in patients who had lymph node metastasis (χ2=12.079, P=0.001). However, neither KDM2B nor EZH2 expression was associated with patients’ age. These results demonstrated that KDM2B and EZH2 positivity in ovarian carcinoma was significantly associated with the tumor histological type, tumor grade, FIGO stage, and lymph node metastasis but not with patients’ age.
KDM2B knockdown decreased EZH2 expression in A2780 and SKOV3 ovarian cancer cells
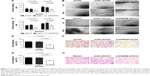
To investigate the effect of KDM2B on EZH2 expression in A2780 and SKOV3 cells, we selectively silenced the KDM2B gene via the lentiviral expression vector of KDM2B shRNA. The inhibition efficiency of KDM2B and the expression of EZH2 were subsequently detected by RT-qPCR and Western blot analysis. The lentiviral shNC expression vector was regarded as a negative control, and cells that were not transduced with the lentiviral expression vector were regarded as a blank control. KDM2B mRNA in shKDM2B-A2780 was reduced by 81.93%±4.56% compared with untransduced A2780 cells (P<0.001), while KDM2B mRNA levels of shNC-A2780 and untransduced cells were not significantly different (P>0.05). In shKDM2B-SKOV3 cells, the KDM2B mRNA level was reduced by 70.66%±6.19%, which was lower than that in shNC-SKOV3 or untransduced cells (P<0.001; Figure 2A). There were no significant differences between negative and blank controls (P>0.05). Furthermore, we found that the EZH2 mRNA levels were decreased by 42.31%±5.76% in shKDM2B-A2780 cells and 32.65%±6.52% in shKDM2B-SKOV3 cells after KDM2B knockdown (P<0.001; Figure 2B).
KDM2B and EZH2 protein levels in A2780 and SKOV3 cells were further confirmed by Western blot analysis. Similarly, compared with untransduced A2780 and SKOV3 cells, KDM2B protein levels were decreased by 35.01%±9.96% and 26.11%±5.17% in shKDM2B-A2780 and shKDM2B-SKOV3 cells, respectively (P=0.005 and 0.001; Figure 2C and D). Meanwhile, the EZH2 protein levels were decreased by 23.79%±4.07% and 21.81%±4.36% in shKDM2B-A2780 and SKOV3 cells compared with those in shNC-A2780 and shNC-SKOV3 or untransduced A2780 and SKOV3 cells (P=0.005 and 0.001; Figure 2E).
KDM2B silencing reduced proliferation capability of ovarian cancer cell lines
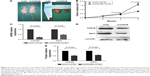
To evaluate the effect of KDM2B knockdown on cell proliferation, CCK-8 assay was performed. Based on the results of the CCK-8 assay, the cell growth curves demonstrated that the proliferation ability of shKDM2B-A2780 and shKDM2B-SKOV3 cells was significantly reduced on days 2, 3, 4, and 5 compared with that of the shNC-A2780, shNC-SKOV3, and untransduced cells. These differences were statistically significant (P<0.001; Figure 3A and B). Our results indicated that the proliferation capability of ovarian cancer was suppressed by KDM2B downregulation.
We speculated that KDM2B was involved in ovarian cancer proliferation through cell cycle regulation. To confirm our hypothesis, we further performed cell cycle analysis using flow cytometry. The result showed that KDM2B depletion induced a slight G1 arrest in ovarian cancer cells, indicating that KDM2B may contribute to cell cycle progression from phase G1 to S (Figure 3C–E). shKDM2B-A2780 cells were characterized by a slight but significant increase of the percentage of cells in phase G0/G1 (P<0.05), while the percentage of cells in the S and G2/M phases was reduced (P<0.05). These data revealed that KDM2B was a cell cycle regulator that participated in controlling the proliferation capability of ovarian cancer cells.
KDM2B deficiency repressed cell migration in ovarian cancer cells
The role of KDM2B in ovarian cancer cell migration was subsequently analyzed by wound healing assay and transwell migration assay. Wound healing assay revealed that the scratch healing rates of shKDM2B-A2780 cells were 14.12%±4.60% at 24 h and 22.72%±4.83% at 48 h, which were low compared with those of shNC-A2780 and untransduced A2780 cells at the same time points (P=0.002, P<0.001; Figure 4A and B). For shKDM2B-SKOV3 cells, the scratch healing rates at 24 h and 48 h were 9.94%±2.64% and 23.30%±4.68%, respectively, which were lower than those of shNC-SKOV3 and untransduced SKOV3 cells at the same time points (P<0.001, P<0.001; Figure 4C and D).
Transwell migration assay produced similar results. Compared with shNC-A2780 and shNC-SKOV3 cells, the migration ability of shKDM2B-A2780 and shKDM2B-SKOV3 cells to cross an 8 μm pore size polycarbonate membrane was significantly inhibited when KDM2B was knocked down (P<0.001, P<0.001; Figure 4E–H). There was no significant difference between untransduced A2780 cells and shNC-A2780 cells (P>0.05), as well as untransduced SKOV3 cells and shNC-SKOV3 cells (P>0.05). The results demonstrated that KDM2B knockdown could repress ovarian cancer cell migration in vitro.
KDM2B silencing inhibited ovarian tumorigenesis in vivo
To further investigate the function of KDM2B on ovarian tumorigenesis, we employed a nude mouse xenograft model. We focused our attention on the A2780 cell line, which easily allowed stable transfection. shKDM2B-A2780 or shNC-A2780 cells were inoculated into the dorsal flank of five nude mice in each group. The tumor sizes of each group were measured every week, and we found that KDM2B knockdown inhibited tumor growth in nude mice after 4 weeks (P<0.001; Figure 5A and B). The RT-qPCR and Western blot results indicated that the EZH2 mRNA and protein level decreased in shKDM2B-A2780 cells compared with shNC-A2780 cells. There was a significant difference between the two groups (P<0.001, P<0.001; Figure 5C–E). These results demonstrated that KDM2B depletion could inhibit ovarian tumorigenesis by EZH2 suppression.
Discussion
Ovarian carcinoma, a malignant genital tumor, threatens the health and life of many women due to its poor prognosis. Although studies on ovarian cancer are increasingly performed, its precise molecular mechanism is still not fully understood. Carcinogenesis depends on alterations of multiple genes, steps, and stages involving oncogene activation and tumor suppressor inactivation. Epigenetic regulation comprises DNA methylation, histone modifications, chromatin remodeling, and non-coding RNA regulation, which plays an important role in many physical or pathological processes, such as cell growth, cell apoptosis, drug resistance, and the self-renewal of stem cells.20–23
Multiple studies have indicated that epigenetic alteration may be an important molecular mechanism contributing to the development and progression of many malignant tumors. Villalba et al observed an increased level of DNA methylation in oncogene promoter regions in lung cancer tissue.24 Abnormal expression of microRNAs has been reported in gastric cancer,25 breast cancer,26 and glioblastoma,27 and this is associated with a poor prognosis.
Histone demethylase KDM2B belongs to the JHDM family. The protein encoded by the KDM2B gene contains JmjC domain, CXXC motif, PHD domain, proline-rich region, and leucine-rich repeat. Among them, the JmjC domain is the active center that can catalyze the demethylation of H3K36me2 and H3K4me3.28–31 Recently, some studies have suggested that KDM2B regulates gene expression in a JmjC-dependent manner and may be involved in tumorigenesis; thus, it has gradually received increasing research attention.25,32 KDM2B overexpression in mouse embryonic fibroblasts (MEFs) induces cellular proliferation and inhibits apoptosis, leading to the immortalization of normal cells.7
Our study demonstrated for the first time that the expression level of KDM2B in normal ovarian tissue, benign tumor, borderline tumor, and ovarian carcinoma was gradually increased and closely associated with the histological type, tumor grade, FIGO stage, and lymph node metastasis. Moreover, KDM2B gene silencing using lentiviral vectors suppressed the proliferation and migration of ovarian cancer cells in vitro and in vivo. These results demonstrated that KDM2B plays an important role in the development and progression of ovarian cancer and could be regarded as a genetic marker of tumor malignancy.
Kottakis et al found that RNA interference in KDM2B reduced the numbers of cell colonies and inhibited cell cycle progression in breast cancer.8 Moreover, Tzatsos et al discovered that KDM2B was highly expressed in pancreatic cancer compared with normal pancreatic tissue and was associated with the poorly differentiated type and aggressive disease.33 These findings are in agreement with our results. However, other researchers reported suppression of KDM2B in certain tumors. Frescas et al confirmed that the expression of KDM2B in human glioblastoma was low.34 They also observed that KDM2B could disturb the proliferation and metabolism of tumor cells by repressing the transcription of ribosomal RNA genes and concluded that KDM2B acts as a tumor suppressor gene in human glioblastoma. Taking these results together, we speculate that the biological functions of KDM2B are distinct in different histological types of tumors, and the exact mechanism needs to be studied in the future.
EZH2 is another key epigenetic regulator that represses transcription by promoting methylation of H3K27 in target genes. Many studies have shown that EZH2 overexpression can be detected in multiple malignant tumors, including prostate cancer,35 breast cancer,36 cervical cancer,37 pancreatic cancer,38 gastric cancer,39 and bladder cancer,40 and that it behaves as an oncogene during carcinogenesis. In our previous research, we found an increased level of EZH2 in ovarian cancer tissue, while the proliferation ability of ovarian cancer cells was significantly restrained when EZH2 was depleted.18 Although there has been plenty of research on the roles of EZH2 in cancer formation, how EZH2 is regulated is rarely discussed. Ma et al discovered that microRNA-26a decreased the expression of EZH2 in hepatoma cells.41 Moreover, Yang et al confirmed that cyclin E and cyclin-dependent kinase-2 could promote EZH2 activity by enhancing its phosphorylation after protein translation.42
Tzatsos et al found that depletion of KDM2B in MEFs leads to EZH2 reduction.43 In ovarian cancer, however, their relationships have not been studied previously. Our research indicates that EZH2 expression is gradually increased with increasing levels of ovarian tumor malignancy and is associated with histological type, tumor grade, FIGO stage, and lymph node metastasis in malignant tissue. Furthermore, KDM2B expression is positively correlated with EZH2. Repressing the expression of KDM2B in ovarian cancer cells significantly lessened the amount of EZH2. This suggests that EZH2 may be one of the target genes affected by the epigenetic regulation of KDM2B. Therefore, in the progression of ovarian tumor, high KDM2B expression may indirectly increase the level of H3K27me3 in the promoter regions of cancer-related genes by regulating EZH2, thereby influencing the transcription of these target genes and contributing to tumorigenesis. Our results preliminarily illustrated the regulatory relationship between KDM2B and EZH2 in ovarian cancer. However, the specific mechanisms by which KDM2B regulates EZH2 need to be further explored.
Conclusion
The increased expression of KDM2B in ovarian cancer was associated with histological type, tumor grade, FIGO stage, and lymph node metastasis, and it was positively correlated with EZH2. KDM2B depletion reduced the expression of EZH2 in ovarian cancer and suppressed the capability of cell proliferation and migration in vitro. Our study confirmed the role of KDM2B in EZH2 epigenetic regulation in ovarian cancer for the first time, preliminarily revealing that certain histone methyltransferase is positively regulated by certain histone demethylase in the epigenetic regulation of ovarian tumors. KDM2B may be a potential genetic marker for ovarian cancer diagnosis and a novel molecular target for clinical therapy.
Acknowledgments
This study was funded by National Natural Science Foundation of China (No 81360389), the National Natural Science Foundation of Guangxi (2012GXNSFAA053086), and the Self-financing Project of the Guangxi Health Department (Z2013026). Yan Kuang and Fangfang Lu are co-first authors for this study.
Disclosure
The authors report no conflicts of interest in this work.
References
Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457–463. | ||
Iacobuzio-Donahue CA. Epigenetic changes in cancer. Annu Rev Pathol. 2009;4:229–249. | ||
Hassler MR, Egger G. Epigenomics of cancer–emerging new concepts. Biochimie. 2012;94(11):2219–2230. | ||
Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8(4):286–298. | ||
Biancotto C, Frigè G, Minucci S. Histone modification therapy of cancer. Adv Genet. 2010;70:341–386. | ||
Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25(1):1–14. | ||
Tzatsos A, Pfau R, Kampranis SC, Tsichlis PN. Ndy1/KDM2B immortalizes mouse embryonic fibroblasts by repressing the Ink4a/Arf locus. Proc Natl Acad Sci U S A. 2009;106(8):2641–2646. | ||
Kottakis F, Foltopoulou P, Sanidas I, et al. NDY1/KDM2B functions as a master regulator of polycomb complexes and controls self-renewal of breast cancer stem cells. Cancer Res. 2014;74(14):3935–3946. | ||
Pfau R, Tzatsos A, Kampranis SC, Serebrennikova OB, Bear SE, Tsichlis PN. Members of a family of JmjC domain-containing oncoproteins immortalize embryonic fibroblasts via a JmjC domain-dependent process. Proc Natl Acad Sci U S A. 2008;105(6):1907–1912. | ||
Yu X, Wang J, Wu J, Shi Y. A systematic study of the cellular metabolic regulation of Jhdm1b in tumor cells. Mol Biosyst. 2015;11(7):1867–1875. | ||
Ren Y, Wu L, Li X, Li W, Yang Y, Zhang M. FBXL10 contributes to the progression of nasopharyngeal carcinoma via involving in PI3K/mTOR pathway. Neoplasma. 2015;62(6):925–931. | ||
Yamaguchi H, Hung MC. Regulation and role of EZH2 in cancer. Cancer Res Treat. 2014;46(3):209–222. | ||
Jia N, Li Q, Tao X, Wang J, Hua K, Feng W. Enhancer of zeste homolog 2 is involved in the proliferation of endometrial carcinoma. Oncol Lett. 2014;8(5):2049–2054. | ||
Zhang Q, Padi SK, Tindall DJ, Guo B. Polycomb protein EZH2 suppresses apoptosis by silencing the proapoptotic miR-31. Cell Death Dis. 2014;5:e1486. | ||
Lin C, Huang F, Li QZ, Zhang YJ. miR-101 suppresses tumor proliferation and migration, and induces apoptosis by targeting EZH2 in esophageal cancer cells. Int J Clin Exp Pathol. 2014;7(10):6543–6550. | ||
Debeb BG, Gong Y, Atkinson RL, et al. EZH2 expression correlates with locoregional recurrence after radiation in inflammatory breast cancer. J Exp Clin Cancer Res. 2014;33(1):58. | ||
Garipov A, Li H, Bitler BG, Thapa RJ, Balachandran S, Zhang R. NF-YA underlies EZH2 upregulation and is essential for proliferation of human epithelial ovarian cancer cells. Mol Cancer Res. 2013;11(4):360–369. | ||
Guo J, Cai J, Yu L, Tang H, Chen C, Wang Z. EZH2 regulates expression of p57 and contributes to progression of ovarian cancer in vitro and in vivo. Cancer Sci. 2011;102(3):530–539. | ||
Karoopongse E, Yeung C, Byon J, et al. The KDM2B-let-7b -EZH2 axis in myelodysplastic syndromes as a target for combined epigenetic therapy. PLoS One. 2014;9(9):e107817. | ||
Zhang F, Xu L, Xu L, et al. JMJD3 promotes chondrocyte proliferation and hypertrophy during endochondral bone formation in mice. J Mol Cell Biol. 2015;7(1):23–34. | ||
Haag T, Herkt CE, Walesch SK, Richter AM, Dammann RH. The apoptosis associated tyrosine kinase gene is frequently hypermethylated in human cancer and is regulated by epigenetic mechanisms. Genes Cancer. 2014;5(9–10):365–374. | ||
Hasemann MS, Lauridsen FK, Waage J, et al. C/EBPα is required for long-term self-renewal and lineage priming of hematopoietic stem cells and for the maintenance of epigenetic configurations in multipotent progenitors. PLoS Genet. 2014;10(1):e1004079. | ||
Zhang YW, Zheng Y, Wang JZ, et al. Integrated analysis of DNA methylation and mRNA expression profiling reveals candidate genes associated with cisplatin resistance in non-small cell lung cancer. Epigenetics. 2014;9(6):896–909. | ||
Villalba M, Diaz-Lagares A, Redrado M, et al. Epigenetic alterations leading to TMPRSS4 promoter hypomethylation and protein overexpression predict poor prognosis in squamous lung cancer patients. Oncotarget. 2016;7(16):22752–22769. | ||
Hong X, Xu Y, Qiu X, et al. MiR-448 promotes glycolytic metabolism of gastric cancer by downregulating KDM2B. Oncotarget. 2016;7(16):22092–22102. | ||
Zehentmayr F, Hauser-Kronberger C, Zellinger B, et al. Hsa-miR-375 is a predictor of local control in early stage breast cancer. Clin Epigenetics. 2016;8:28. | ||
Ayala-Ortega E, Arzate-Mejía R, Pérez-Molina R, et al. Epigenetic silencing of miR-181c by DNA methylation in glioblastoma cell lines. BMC Cancer. 2016;16:226. | ||
Polytarchou C, Pfau R, Hatziapostolou M, Tsichlis PN. The JmjC domain histone demethylase Ndy1 regulates redox homeostasis and protects cells from oxidative stress. Mol Cell Biol. 2008;28(24):7451–7464. | ||
Kavi HH, Birchler JA. Drosophila KDM2 is a H3K4me3 demethylase regulating nucleolar organization. BMC Res Notes. 2009;2:217. | ||
Inagaki T, Iwasaki S, Matsumura Y, et al. The FBXL10/KDM2B scaffolding protein associates with novel polycomb repressive complex-1 to regulate adipogenesis. J Biol Chem. 2015;290(7):4163–4177. | ||
Salminen A, Kaarniranta K, Kauppinen A. Hypoxia-inducible histone lysine demethylases: impact on the aging process and age-related diseases. Aging Dis. 2016;7(2):180–200. | ||
Ueda T, Nagamachi A, Takubo K, et al. Fbxl10 overexpression in murine hematopoietic stem cells induces leukemia involving metabolic activation and upregulation of Nsg2. Blood. 2015;125(22):3437–3446. | ||
Tzatsos A, Paskaleva P, Ferrari F, et al. KDM2B promotes pancreatic cancer via Polycomb-dependent and -independent transcriptional programs. J Clin Invest. 2013;123(2):727–739. | ||
Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450(7167):309–313. | ||
Chinaranagari S, Sharma P, Chaudhary J. EZH2 dependent H3K27me3 is involved in epigenetic silencing of ID4 in prostate cancer. Oncotarget. 2014;5(16):7172–7182. | ||
Guo S, Li X, Rohr J, et al. EZH2 overexpression in different immunophenotypes of breast carcinoma and association with clinicopathologic features. Diagn Pathol. 2016;11(1):41. | ||
Chen Q, Zheng PS, Yang WT. EZH2-mediated repression of GSK-3β and TP53 promotes Wnt/β-catenin signaling-dependent cell expansion in cervical carcinoma. Oncotarget. 2016;7(24):36115–36129. | ||
Han T, Jiao F, Hu H, et al. EZH2 promotes cell migration and invasion but not alters cell proliferation by suppressing E-cadherin, partly through association with MALAT-1 in pancreatic cancer. Oncotarget. 2016;7(10):11194–11207. | ||
Chen WM, Huang MD, Sun DP, et al. Long intergenic non-coding RNA 00152 promotes tumor cell cycle progression by binding to EZH2 and repressing p15 and p21 in gastric cancer. Oncotarget. 2016;7(9):9773–9787. | ||
Zhang Q, Zhao W, Ye C, et al. Honokiol inhibits bladder tumor growth by suppressing EZH2/miR-143 axis. Oncotarget. 2015;6(35):37335–37348. | ||
Ma DN, Chai ZT, Zhu XD, et al. MicroRNA-26a suppresses epithelial-mesenchymal transition in human hepatocellular carcinoma by repressing enhancer of zeste homolog 2. J Hematol Oncol. 2016;9:1. | ||
Yang CC, LaBaff A, Wei Y, et al. Phosphorylation of EZH2 at T416 by CDK2 contributes to the malignancy of triple negative breast cancers. Am J Transl Res. 2015;7(6):1009–1020. | ||
Tzatsos A, Paskaleva P, Lymperi S, et al. Lysine-specific demethylase 2B (KDM2B)-let-7-enhancer of zester homolog 2 (EZH2) pathway regulates cell cycle progression and senescence in primary cells. J Biol Chem. 2011;286(38):33061–33069. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.