Back to Journals » OncoTargets and Therapy » Volume 12
Histone deacetylase 6 is overexpressed and promotes tumor growth of colon cancer through regulation of the MAPK/ERK signal pathway
Authors Zhang SL, Zhu HY, Zhou BY, Chu Y , Huo JR, Tan YY, Liu DL
Received 17 November 2018
Accepted for publication 1 March 2019
Published 2 April 2019 Volume 2019:12 Pages 2409—2419
DOI https://doi.org/10.2147/OTT.S194986
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Carlos E Vigil
Shi-Lan Zhang, Hong-Yi Zhu, Bing-Yi Zhou, Yi Chu, Ji-Rong Huo, Yu-Yong Tan, De-Liang Liu
Department of Gastroenterology, The Second Xiangya Hospital, Central South University, Changsha, Hunan 410011, People’s Republic of China
Purpose: To investigate the expression of histone deacetylase 6 (HDAC6) in colon cancer and its role in colon cancer cell growth and migration.
Materials and methods: We detected the expression of HDAC6 in a colon cancer tissue chip using immunochemical staining, and analyzed the difference in HDAC6 expression between cancer and adjacent noncancerous tissues. Then, we explored the relationship between HDAC6 expression and patients’ clinicopathological characteristics and prognoses. In adidition, the role of HDAC6 in colon cancer cell growth and migration, as well as its potential related signal pathway, through HDAC6 knockdown was explored.
Results: The immunochemical score of HDAC6 expression was higher in cancer tissue than in the adjacent noncancerous tissue (4.54 vs 3.08, P<0.005); similarly, as well as the rate of high HDAC6 expression was higher in cancer tissue than in the adjacent noncancerous tissue (71.1% vs 40.9%, P<0.001). Patients showing high HDAC6 expression had a shorter overall survival time. Additionally, Cox regression analysis showed that high HDAC6 expression was an independent risk factor for poor prognosis. HDAC6 knockdown decreased cell viability, colony formation, and number of migrated colon cancer cells (HCT116 and HT29); the expression of p-MEK, p-ERK, and p-AKT was also decreased, but had no influence on MEK, ERK, and AKT expression.
Conclusion: HDAC6 is highly expressed in colon cancer and associated with a poor prognosis. HDAC6 knockdown inhibits colon cancer cell growth and migration, partly through the MAPK/ERK pathway.
Keywords: colon cancer, cell growth and migration, histone deacetylase 6, MAPK/ERK pathway, CRC prognosis
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide, ranked third in cancer incidence and second in mortality.1–3 Although surgery, chemotherapy, radiotherapy, and biological target therapy have been used in the treatment of colorectal cancer and have greatly improved the prognosis of CRC patients, even today over 50% of patients die due to tumor recurrence or metastasis.4 What’s more, the underlying mechanism of the disease remains largely undefined. Therefore, further research is required to explore the pathogenic mechanism of CRC and new therapeutic strategies to combat it.
Acetylation and deacetylation play important roles in tumorigenesis, and deacetylation is mediated by histone deacetylases (HDACs). HDACs alter the acetylation status of histone and non-histone proteins to regulate various cellular events, such as proliferation, differentiation, apoptosis, autophagy, angiogenesis and tumor immunity, in tumor cells. HDACs can be divided into four classes based on their homology to yeast HDACs: class I, class II, class III and class VI. In addition, class II can be further divided into IIa and IIb.5,6
Histone deacetylase 6 (HDAC6) is a unique Class IIb HDACs, in that it is a predominant cytoplasmic protein with two deacetylase domains, and it has been demonstrated to promote tumor growth in many human cancers including gastrointestinal cancers.7,8 In our previous study, we found that HDAC6 knockdown could suppress proliferation, migration, and invasion in HCT116 cells.9 Other research has found that HDAC6-selective inhibitors could inhibit colon cancer cell growth, proliferation and, invasion.10–15 However, the expression of HDAC6 in colon cancer tissue and its relationship with patients’ clinicopathologic characteristics remain largely unknown.
In the present study, we first examined the expression of HDAC6 in clinical colon cancer patients and explored the association between HDAC6 expression and patients’ clinical features. Then we investigated the effects of HDAC6 on the cellular biological behavior of colon cancer cells and presented evidence of HDAC6 interacting with the MAPK/ERK signaling pathway. The results of this study may provide a better understanding of the role HDAC6 plays in colon cancer, as well as a rationale for clinical investigations or for the application of HDAC6-selective inhibitors in CRC patients.
Materials and methods
Immunohistochemistry
Human tissue microarrays were purchased from Shanghai Outdo Biotech, (catalog no. HCol-Ade180Sur-07, Shanghai, China). Demographic and clinicopathological data, including clinical stage (according to the American Joint Committee on Cancer staging system) and survival data were provided by the manufacturer. Tissue sections were deparaffinized, soaked in 0.01 M sodium citrate buffer and boiled in a microwave oven for 5 min at 500 W to retrieve antigens. After treatment with 3% hydrogen peroxide to inactivate endogenous peroxidase and blocking with normal goat serum to reduce non-specific staining, the sections were incubated with anti-HDAC6 antibody (7558; Cell Signaling Technology, USA) at a dilution of 1:400 overnight at 4 °C. Tissue sections were immunohistochemically stained using Envision reagents (Envision+Dual link/HRP; Dako Denmark). All sections were counterstained with hematoxylin. The immunohistochemical (IHC) score was assessed with the Fromowitz standard:16 0–5%=0; 5–25%=1; 26–50%=2; 51–75%=3; >75%=4. The staining intensity was graded as follows: absent or faint blush=0; weak=1; moderate=2; strong=3. The final IHC score was calculated by adding the percentage and intensity scores. Thus, the minimum score was 0, the maximum score was 7, and the grading was as follows: 0–1=(−), 2–3=(+), 4–5=(++), 6–7=(+++). A final score ≤3 was considered low expression, while and ≥4 was considered high expression. Two experienced pathologists, blinded to all clinicopathological information on the patients, independently evaluated the immunostaining scores. The cases eliciting disagreement were reevaluated jointly for consensus.
Cell lines and cell culture
Human colon cancer HCT116 and HT29 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in DMEM medium (Life Technologies, CA) containing 10% fetal bovine serum (FBS), 100 units of penicillin and 100 mg/ml streptomycin. The cells were maintained in a 5% CO2 culture incubator at 37 °C.
Reagents and antibodies
Anti-HDAC6 (7558), anti-phospho-s473-AKT (9018), anti-AKT (2938), anti-phospho-MEK (9154), anti-MEK (8727), anti-phospho-ERK (3192), anti-ERK (4695) and anti-Ac-α-Tubulin (5335) antibodies were purchased from Cell Signaling Technology. Anti-Tubulin (SC-73242) antibody was purchased from Santa Cruz. All antibodies were used in 1:1,000 dilutions in 5% non-fat milk for western blot.
Plasmids
ShScramble, shHDAC6-1, and shHDAC6-2 were described previously.9 Lipofectamine 2,000 Transfection Reagent (Thermo Fisher Scientific, USA) was used for transfection according to the protocol recommended by the manufacturer. A 293FT cell line was to package and amplify the lentivirus. The media containing viruses were collected 48 hrs and 72 hrs after transfection. After filtering through 0.45 µM filters, viruses were used to infect cells in the presence of 4 μg/mL polybrene (Sigma-Aldrich). At 48 hrs post-infection, cells were split and selected using 1 μg/mL puromycin (Sigma-Aldrich) for three days to eliminate the uninfected cells before harvesting.
Cell viability assay
Cell treated with sh-Scramble, shHDAC6-A, and shHDAC6-B were respectively plated at 3,000 cells/well in 96-well plates and cultured in a 5% CO2 culture incubator at 37 °C. After 24 hrs, 48 hrs, 72 hrs, and 96 hrs, cell viability was detected with the Cell Titer-Glo Luminescent Cell Viability Assay kit according to the manufacturer’s instructions (Promega).
Colony formation assays
HCT116 and HT29 cells treated with sh-Scramble, shHDAC6-A, and shHDAC6-B were respectively plated at a density of 1,000 cells/well in 6-well culture dishes and allowed to grow undisturbed for 10 days. Cells were stained with crystal violet and the colony numbers were counted.
Transwell assay
HCT116 cells and HT29 cells treated with sh-Scramble, shHDAC6-A, and shHDAC6-B were respectively plated at a density of 5×104 cells in an 8-μm, 24-well plate chamber insert (Corning Life Sciences, catalog no. 3422), with 100 μL serum-free medium at the top of the insert and DMEM medium (Gibco) containing 10% FBS (500 μL) at the bottom. Cells were incubated for 24 h and fixed with 4% paraformaldehyde for 15 min. After washing with PBS, cells at the top of the insert were scraped with a cotton swab. Cells adhering to the bottom were stained with 0.5% crystal violet blue for 15 min and then washed with double-distilled H2O. The positively stained cells were counted in eight random fields under the microscope, and the average value of the eight fields was expressed. All assays were performed in triplicate.
Western blot assay
Cells treated with sh-Scramble, shHDAC6-A, and shHDAC6-B were harvested and washed once using cold PBS. The cell pellets were lysed in EBC buffer (50 mM Tris pH 7.5, 120 mM NaCl, 0.5% NP40) supplemented with protease inhibitors (Roche) and phosphatase inhibitors (Calbio chem). The lysates were cleared by centrifugation and then quantified with a Beckman Coulter DU-800 spectrophotometer (Beckman Coulter) using the Bio-Rad protein assay reagent (Bio-Rad Laboratories, CA). The lysates were eluted by boiling for 5 mins in SDS loading buffer. Bound proteins were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
Statistical analysis
Data were analyzed by with the SPSS 23.0 software (Chicago, IL, USA). The data were expressed as the mean and SD and compared by ANOVA unless stated otherwise. HDAC6 expression in tumor tissues and matched non-tumor tissues was analyzed with the paired Student’s t-test. The association between HDAC6 expression and various clinicopathological parameters was evaluated with the Chi-Square test. The Cox proportional hazard regression model was used for univariate and multivariate analyses to determine the effects of the clinicopathological variables and HDAC6 expression on patient survival. Only variables with P-values <0.1 in the univariate analysis were included in the multivariate analysis.
Results
HDAC6 was overexpressed in colon cancer tissues and associated with poor prognosis
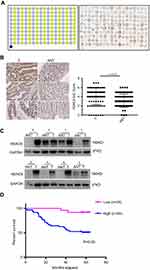
Immunohistochemical staining was performed on a tissue microarray containing 180 cores/spots consisting of paired colon cancer tissue and corresponding adjacent non-neoplastic mucosa tissue from 90 patients (Figure 1A). Among the 90 pairs, 7 pairs were excluded due to a lack of either detectable tumor cells or normal colon epithelial cells, so that a total of 83 pairs were enrolled for analysis. As illustrated in Figure 1B, HDAC6 was expressed in both the cytoplasm and nucleus (but mostly in the cytoplasm), and cytoplasmic HDAC6 staining was evaluated in the study. Our results showed that the IHC score of HDAC6 expression in the cancer tissue was higher than in the matching noncancerous tissue (4.54 vs 3.08, P<0.005) (Figure 1B, right). Out of the analyzed cases of colon cancer, 71.1% (59/83) showed strong cytoplasmic HDAC6 staining, which was significantly higher than that in noncancerous tissue (40.9%, 34/83). We then explored the correlation between HDAC6 expression and the clinicopathological characteristics of colon cancer patients. The data showed that the HDAC6 expression levels were associated with tumor size, but not with other parameters such as age, gender, tumor site, lymph node invasion, differentiation grade, JACC clinical stage, etc (Table 1). Univariate analysis revealed that tumor size, lymph node invasion, JACC clinical stage, vascular invasion, and high HDAC6 expression were significantly associated with patients’ survival (Table 2). Furthermore, multivariate Cox regression analysis confirmed lymph node invasion, vascular invasion, and high HDAC6 expression as independent risk factors for poor survival (Table 2). Kaplan-Meier analysis demonstrated that those patients with high HDAC6 expression had a shorter overall survival time (median=55 months) compared to patients with low HDAC6 expression (median=58 months, P<0.05, Figure 1D). We also detected HDAC6 expression in 8 colon cancer pairs using western blot, and the results showed that HDAC6 was up-regulated compared with adjacent noncancerous tissue (Figure 1C).
 | Table 1 Relationship between HDAC6 expression and clinicopathologic features of CRC patients |
 | Table 2 Univariable and multivariable analyses of OS and clinicopathologic variables in CRC |
HDAC6 knockdown inhibits colon cancer cell proliferation, migration, and invasion
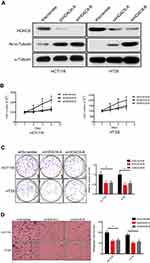
Since HDAC6 was overexpressed in colon cancer and correlated with poor prognosis, we next determined whether HDAC6 played an oncogenic role in colon cancer. We investigated the biological effects of HDAC6 knockdown in two colon cancer cell lines (HCT116 and HT29) using shRNA (Figure 2A). The results showed that HDAC6 knockdown significantly decreased cell viability and the colony formation number, suggesting cell growth inhibition by HDAC6 knockdown (Figure 2B–C)
Cell migration and invasion are essential to cancer metastasis.17 Thus, we further investigated the potential impact of HDAC6 knockdown on the metastatic properties of colon cancer cells using transwell assays, and found that HDAC6 knockdown with either of shRNA significantly impaired cell migration potential (Figure 2D). The results demonstrated that HDAC6 suppression may inhibit colon cancer metastasis and invasion.
HDAC6 exert its function partly through the MAPK/ERK signaling pathway
Mitogen-activated protein kinases (MAPKs) consisting of ERK, JNK and p38 are a major class of serine-threonine kinases that form the main cell proliferation and migration signaling pathway from the cell surface to the nucleus.18,19 MAPK/ERK signaling pathways play critical roles in tumor proliferation, migration, and invasion.20 A growing body of evidence indicated that activation of the MAPK/ERK pathway promotes CRC progression and inhibition of this pathway may be used to treat CRC.20,21
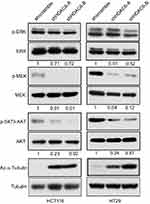
We therefore determined whether HDAC6 knockdown had an effect on the MAPK/ERK signaling pathway, and the results showed that HDAC6 knockdown could decrease the expression of p-MEK, p-ERK and p-AKT, but had no obvious effect on MEK, ERK and, AKT expression in either cell line (Figure 3). These data suggest that HDAC6 knockdown exerts its function partly through the MAPK/ERK signaling pathway in colon cancer cells.
Discussion
In the present study, we found that HDAC6 is highly expressed in colon cancer, and its high expression is associated with poor prognosis. To the best of our knowledge, this is the first study reporting on the expression levels of HDAC6 in colon cancer, as well as on its association with patients’ clinical characteristics. We further certified the oncogenic role of HDAC6 in colon cancer via cell experiments. The results of the present study help to better understand the mechanism of colon cancer development, and provide a rational basis for the clinical application of HDAC6-selective inhibitors in the treatment of colon cancer.
HDAC6 is a unique Class IIb histone deacetylase that plays vital roles in many human cancers. In gastrointestinal cancers, the role of HDAC6 varies with the cancer type and tumor microenvironment. For example, HDAC6 is an oncogene in esophageal cancer, while it can both an oncogene and a tumor suppressor in hepatocellular and gastric cancer.22–24 Our study showed that HDAC6 was highly expressed in colon cancer tissues, and a high expression of HDAC6 predicted a shorter overall survival. Furthermore, HDAC6 knockdown could inhibit tumor growth and migration. Several previous studies have shown that HDAC6 inhibitors can inhibit the growth of colon cancer cell lines (HCT116, HT29, Caco-2).10,11,13,25,26 However, more recent research reported that a HDAC inhibitor (TriA) showed no effect on migration in colon cancer cells induced by TPA (a tumor promoter). Differently from previous studies, this latest study focused on the extent to which gene promoter methylation induced by reactive oxygen species facilitated the migration and invasion of colon cancer cells but not on the effect of histone deacetylases, which are two different mechanisms of tumorigenesis. Overall, the results indicated that methylation was also one of the factors responsible for colon cancer metastasis, which may serve as a therapeutic tool in the future.27
Previous studies have demonstrated that HDAC6 plays an important role in various cellular process, such as proliferation, differentiation, apoptosis, autophagy, and angiogenesis, and in the underlying signal pathways involved, including MAPK/ERK, TGF-β, mTOR, NF-κB, etc.28–31 Since HDAC6-selective inhibitors showed a suppressive effect on colon cancer cells via the MARP/ERK signal pathway, and since the MAPK/ERK pathway plays an important role in colon cancer development,32,33 we investigated the role of HDAC6 knockdown on the MAPK/ERK pathway in colon cancer cell lines. As the results showed, HDAC6 knockdown significantly decreased the levels of p-ERK, p-MEK and p-AKT, while no obvious change was noticed in MEK, ERK and AKT expression, suggesting that HDAC6 exerts its function partly through the MAPK/ERK signal pathway, which is consistent with the results obtained with HDAC6-selective inhibitors.
Additionally, MAPKs are regulated by reactive oxygen species, which were proposed to be involved in tumor metastasis by triggering mitochondrial dysfunction and DNA damage.34 Accumulating evidence suggests that an increase in reactive oxygen species initiates the p38 MAPK signaling pathway and triggers the apoptosis cascade in colon cancer. Therefore, treatment with ROS scavengers or MAPK inhibitors would abolish this effect in cancer cells.35–38 Altogether, measures that target the ROS-induced activation of p38 MAPK provide new insights into the molecular mechanisms that may be of therapeutic interest for the treatment of colon cancer.
Conclusion
In conclusion, our study indicates that HDAC6 plays a critical role in regulating tumor growth and metastasis in colon cancer. This effect maybe partly mediated by the HDAC6-mediated inactivation of MAPK/ERK pathways. Thus, HDAC6 may be a valuable prognostic marker, and targeting HDAC6 may serve as a potential therapeutic approach that could effectively inhibit tumor growth and metastasis in colon cancer patients.
Institutional review board statement
The study was approved by the Ethics Committee of the Second Xiangya Hospital, Central South University, Changsha, China.
Institutional animal care and use committe statement
No animal was used in the present study.
Acknowledgments
This work was supported by Natural Science Foundation of Hunan Province (no. 2019JJ50900) to Yu-Yong Tan and Fundamental Research Funds from the Central Universities of Central South University (no. 2018zzts047) to Shi-Lan Zhang.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Kd M, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
2. Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin. 2012;62(1):10–29. doi:10.3322/caac.20138
3. Saif MW, Chu E. Biology of colorectal cancer. Cancer J. 2010;16(3):196–201. doi:10.1097/PPO.0b013e3181e076af
4. Benson AB
5. Polevoda B, Sherman F. Nalpha-terminal acetylation of eukaryotic proteins. J Biol Chem. 2000;275(47):36479–36482. doi:10.1074/jbc.R000023200
6. Starheim KK, Gromyko D, Velde R, Varhaug JE, Arnesen T. Composition and biological significance of the human Nalpha-terminal acetyltransferases. BMC Proc. 2009;3(Suppl 6):S3. doi:10.1186/1753-6561-3-s6-s3
7. Chhabra S. Novel proteasome inhibitors and histone deacetylase inhibitors: progress in myeloma therapeutics. Pharmaceuticals (Basel). 2017;10:2. doi:10.3390/ph10030067
8. Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science (New York, NY). 2015;347(6220):1260419. doi:10.1126/science.1260419
9. Tan YY, Ci YP, Dai XP, et al. Cullin 3(SPOP) ubiquitin E3 ligase promotes the poly-ubiquitination and degradation of HDAC6. Oncotarget. 2017;8(29):47890–47901. doi:10.18632/oncotarget.18141
10. Gotze S, Coersmeyer M, Muller O, Sievers S. Histone deacetylase inhibitors induce attenuation of Wnt signaling and TCF7L2 depletion in colorectal carcinoma cells. Int J Oncol. 2014;45(4):1715–1723. doi:10.3892/ijo.2014.2550
11. Ryu HW, Lee DH, Shin DH, Kim SH, Kwon SH. Aceroside VIII is a new natural selective HDAC6 inhibitor that synergistically enhances the anticancer activity of HDAC inhibitor in HT29 cells. Planta Med. 2015;81(3):222–227. doi:10.1055/s-0034-1396149
12. Fang LT, Lee S, Choi H, et al. Comprehensive genomic analyses of a metastatic colon cancer to the lung by whole exome sequencing and gene expression analysis. Int J Oncol. 2014;44(1):211–221. doi:10.3892/ijo.2013.2150
13. Yang Z, Wang T, Wang F, et al. Discovery of selective histone deacetylase 6 inhibitors using the quinazoline as the cap for the treatment of cancer. J Med Chem. 2016;59(4):1455–1470. doi:10.1021/acs.jmedchem.5b01342
14. Liu YM, Lee HY, Lai MJ, et al. Pyrimidinedione-mediated selective histone deacetylase 6 inhibitors with antitumor activity in colorectal cancer HCT116 cells. Org Biomol Chem. 2015;13(40):10226–10235. doi:10.1039/C5OB01509J
15. Tan Y, Zhang S, Zhu H, et al. Histone deacetylase 6 selective inhibitor ACY1215 inhibits cell proliferation and enhances the chemotherapeutic effect of 5-fluorouracil in HCT116 cells. Ann Transl Med. 2019;7:1. doi:10.21037/atm.2018.11.48
16. Fromowitz FB, Viola MV, Chao S, et al. ras p21 expression in the progression of breast cancer. Hum Pathol. 1987;18(12):1268–1275.
17. Duff D, Long A. Roles for RACK1 in cancer cell migration and invasion. Cell Signal. 2017;35:250–255. doi:10.1016/j.cellsig.2017.03.005
18. Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Annu Rev Immunol. 2002;20:55–72. doi:10.1146/annurev.immunol.20.091301.131133
19. Sun Y, Liu WZ, Liu T, Feng X, Yang N, Zhou HF. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res. 2015;35(6):600–604. doi:10.3109/10799893.2015.1030412
20. Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6(5):322–327. doi:10.1016/S1470-2045(05)70168-6
21. Martin-Liberal J, Lagares-Tena L, Larkin J. Prospects for MEK inhibitors for treating cancer. Expert Opin Drug Saf. 2014;13(4):483–495. doi:10.1517/14740338.2014.892578
22. Seidel C, Schnekenburger M, Dicato M, Diederich M. Histone deacetylase 6 in health and disease. Epigenomics. 2015;7(1):103–118. doi:10.2217/epi.14.69
23. Yang PH, Zhang L, Zhang YJ, Zhang J, Xu WF. HDAC6: physiological function and its selective inhibitors for cancer treatment. Drug Discov Ther. 2013;7(6):233–242. doi:10.5582/ddt.2013.v7.6.233
24. Aldana-Masangkay GI, Sakamoto KM. The role of HDAC6 in cancer. J Biomed Biotechnol. 2011;2011:875824. doi:10.1155/2011/875824
25. Lee HY, Tsai AC, Chen MC, et al. Azaindolylsulfonamides, with a more selective inhibitory effect on histone deacetylase 6 activity, exhibit antitumor activity in colorectal cancer HCT116 cells. J Med Chem. 2014;57(10):4009–4022. doi:10.1021/jm401899x
26. Chao OS, Chang TC, Di Bella MA, et al. The HDAC6 inhibitor tubacin induces release of CD133(+) extracellular vesicles from cancer cells. J Cell Biochem. 2017;118(12):4414–4424. doi:10.1002/jcb.26095
27. Banskota S, Dahal S, Kwon E, Kim DY, Kim JA. beta-Catenin gene promoter hypermethylation by reactive oxygen species correlates with the migratory and invasive potentials of colon cancer cells. Cell Oncol (Dordr). 2018;41(5):569–580. doi:10.1007/s13402-018-0391-7
28. Williams KA, Zhang M, Xiang S, et al. Extracellular signal-regulated kinase (ERK) phosphorylates histone deacetylase 6 (HDAC6) at serine 1035 to stimulate cell migration. J Biol Chem. 2013;288(46):33156–33170. doi:10.1074/jbc.M113.472506
29. Bae HJ, Jung KH, Eun JW, et al. MicroRNA-221 governs tumor suppressor HDAC6 to potentiate malignant progression of liver cancer. J Hepatol. 2015;63(2):408–419. doi:10.1016/j.jhep.2015.03.019
30. Gu S, Liu Y, Zhu B, et al. Loss of alpha-tubulin acetylation is associated with TGF-beta-induced epithelial-mesenchymal transition. J Biol Chem. 2016;291(10):5396–5405. doi:10.1074/jbc.M115.713123
31. Liu F, Zhao X, Qian Y, Zhang J, Zhang Y, Yin R. MiR-206 inhibits head and neck squamous cell carcinoma cell progression by targeting HDAC6 via PTEN/AKT/mTOR pathway. Biomed Pharmacother. 2017;96:229–237. doi:10.1016/j.biopha.2017.08.145
32. Guo Y, Zhang YN, Yang XJ, et al. Effects of methylglyoxal and glyoxalase I inhibition on breast cancer cells proliferation, invasion, and apoptosis through modulation of MAPKs, MMP9, and Bcl-2. Cancer Biol Ther. 2016;17(2):169–180. doi:10.1080/15384047.2015.1121346
33. Yang NM, Cui H, Han F, et al. Paeoniflorin inhibits human pancreatic cancer cell apoptosis via suppression of MMP-9 and ERK signaling. Oncol Lett. 2016;12(2):1471–1476. doi:10.3892/ol.2016.4761
34. Chen JC, Zhang Y, Jie XM, et al. Ruthenium(II) salicylate complexes inducing ROS-mediated apoptosis by targeting thioredoxin reductase. J Inorg Biochem. 2019;193:112–123. doi:10.1016/j.jinorgbio.2019.01.011
35. Cho YW, Kim EJ, Nyiramana MM, et al. Paroxetine induces apoptosis of human breast cancer MCF-7 cells through Ca(2+)-and p38 MAP kinase-dependent ROS generation. Cancers. 2019;11:1. doi:10.3390/cancers11010064
36. Wang JN, Che Y, Yuan ZY, et al. Acetyl-macrocalin B suppresses tumor growth in esophageal squamous cell carcinoma and exhibits synergistic anti-cancer effects with the Chk1/2 inhibitor AZD7762. Toxicol Appl Pharmacol. 2019;365:71–83. doi:10.1016/j.taap.2019.01.005
37. Dabiri Y, Schmid A, Theobald J, et al. A Ruthenium(II) N-Heterocyclic Carbene (NHC) complex with naphthalimide ligand triggers apoptosis in colorectal cancer cells via activating the ROS-p38 MAPK pathway. Int J Mol Sci. 2018;19:12. doi:10.3390/ijms19123964
38. Schroyer AL, Stimes NW, Abi Saab WF, Chadee DN. MLK3 phosphorylation by ERK1/2 is required for oxidative stress-induced invasion of colorectal cancer cells. Oncogene. 2018;37(8):1031–1040. doi:10.1038/onc.2017.396
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.