Back to Journals » Cancer Management and Research » Volume 10
Histone deacetylase 6 expression in metastatic lymph nodes is a valuable prognostic marker for resected node-positive esophageal squamous cell cancer
Authors Xie X, Luo KJ, Li Y, Ling YH, Zhang SS , Xie XY, Wen J
Received 28 June 2018
Accepted for publication 27 September 2018
Published 8 November 2018 Volume 2018:10 Pages 5451—5460
DOI https://doi.org/10.2147/CMAR.S178575
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Kenan Onel
Xuan Xie,1,2,* Kongjia Luo,3–5,* Yi Li,3,4,6 Yihong Ling,3,4,7 Shuishen Zhang,8 Xiuying Xie,3,4 Jing Wen3,4
1Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, People’s Republic of China; 2Department of Thoracic Surgery, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, People’s Republic of China; 3State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, Guangzhou, People’s Republic of China; 4Guangdong Esophageal Cancer Research Institute, Guangzhou, People’s Republic of China; 5Department of Thoracic Surgery, Sun Yat-sen University Cancer Center, Guangzhou, People’s Republic of China; 6Department of Anesthesiology, Sun Yat-sen University Cancer Center, Guangzhou, People’s Republic of China; 7Department of Pathology, Sun Yat-sen University Cancer Center, Guangzhou, People’s Republic of China; 8Department of Thoracic Surgery, the First Affiliated Hospital, Sun Yat-sen University, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Background: Histone deacetylase 6 (HDAC6) exerts enzymatic deacetylation activity on histones and on non-histone substrates and plays a key role in microtubule dynamics and chaperone activities. In addition, previous studies have demonstrated its role in cancer progression. However, its clinical significance in esophageal squamous cell cancer (ESCC) has not been elucidated. We investigated the correlation of HDAC6 expression and clinical outcome in a group of T3N1–3M0 surgically resected ESCCs.
Methods: Tissue microarrays were conducted on 209 surgically resected T3N1–3M0 ESCC tumors, including 163 pairs of primary tumors (PTs) and their corresponding metastatic lymph nodes (MLNs). Immunohistochemistry was utilized to evaluate HDAC6 protein levels. The relationship between patient outcomes and HDAC6 expression was analyzed statistically.
Results: The level of HDAC6 expression in ESCC MLNs was found to be significantly lower than that in PTs (P<0.001). Patients with lower MLN HDAC6 expression demonstrated improved overall survival (P=0.011) and disease-free survival (P=0.012) than those with higher HDAC6 expression. HDAC6 expression levels in PTs revealed no prognostic significance. Multivariate analysis showed that the MLN HDAC6 expression level was an independent prognostic factor for both overall survival (HR 1.456, P=0.029) and disease-free survival (HR 1.432, P=0.033).
Conclusion: High expression of HDAC6 in MLNs but not in PTs suggests a poor prognosis for patients with resected T3N1–3M0 ESCC. We should take into account the protein expression of MLNs when assessing prognosis in patients with lymph-node involvement.
Keywords: esophageal cancer, HDAC6, biomarker, protein expression, outcomes
Introduction
Esophageal cancer constitutes one of the most severe forms of cancer, with a high annual death rate.1 Esophageal cancer can be divided into adenocarcinoma and squamous cell carcinoma based upon histological origin, with adenocarcinoma being more predominant in western countries and squamous cell carcinoma more predominant in eastern countries.1 The TNM staging system is the most useful way to assess prognosis.2 Patients without distant metastasis are candidates for surgery. Among them, patients without nodal involvement exhibit a relatively improved survival outcome. While multiple treatment modalities have been introduced over the past few decades, patients who are positive for node involvement (N+) typically have poor prognosis.3 To improve these patients’ treatment outcomes, it is important to select those eligible for surgery, perform surgery properly, and prescribe follow-up adjuvant therapy to patients with poor prognosis.
Several studies have focused on the issue of the number of metastatic lymph nodes (MLNs),4 MLN stations,5 MLN ratio (MLN number/examined lymph-node number),6 and skip metastases,7 which all proved to be essential in distinguishing patients with different outcomes, but disparities still persisted. Differences in the extent of lymphadenectomy (two-field vs three-field dissection) may also influence patient outcomes based on different numbers and stations of dissected lymph nodes.8 Molecular profiling has been widely used in precision treatment guidance and outcome prediction in many kinds of cancers,9–11 including esophageal squamous cell cancer (ESCC).12 Nonetheless, very few studies have concentrated on patients with MLNs who are still considered surgical candidates.
Histone deacetylases (HDACs) are enzymes involved in the regulation of multiple processes, including gene expression regulation, protein activity, and deacetylation of histone proteins.13 HDAC6 is unique among the HDAC enzyme family, having two active catalytic domains and a unique physiological function.14,15 In addition to the deacetylation of histones, HDAC6 can exert deacetylase enzymatic activity on non-histone substrates, including Hsp90,16 cortactin,17 peroxiredoxin,18 and prolyl isomerase Pin1,19 hence playing a key role in microtubule dynamics, chaperone activities, and tumor progression.20,21 Its dysregulation relates to many kinds of cancers, with variable effects; high expression of HDAC6 has been shown to be associated with tumor development in hepatocellular cancer,22 pancreatic cancer,23 and glioblastoma,24 while decreased expression has been found to be associated with the suppression of proliferation, migration, or invasion in breast cancer,25 lung cancer,26 and gastric cancer.27 In ESCC, Li et al28 found that downregulation of HDAC6 expression could inhibit cell proliferation and reduce cell migration in vitro. However, the role of HDAC6 in vivo and its prognostic value in ESCC patients have not yet been elucidated.
In the current study, the expression levels of HDAC6 protein in N+ ESCC primary tumors (PTs) and the corresponding levels in MLNs were evaluated. Our results revealed high levels of HDAC6 expression in PT tissues and a positive correlation between MLN HDAC6 expression and poor ESCC patient survival.
Patients and methods
Patient selection
The current study was approved by the Ethics Committee of Sun Yat-sen University Cancer Center and all patients provided written informed consent.
Patients were selected retrospectively from those who had undergone esophagectomy with standard or extended dissection of thoracic and abdominal lymph nodes between July 1997 and December 2004 at the Department of Thoracic Surgery, Sun Yat-sen University Cancer Center. Additional selection criteria included 1) pathological proof of thoracic T3N1–3M0 ESCC according to the eighth edition American Joint Committee on Cancer TNM staging system,2 2) the absence of neoadjuvant or adjuvant therapy, 3) complete surgical resection, 4) and sufficient formalin-fixed and paraffin-embedded PT and MLN samples for tissue microarrays (TMAs).
TMA construction
TMAs were constructed using a Beecher Instruments tissue microarrayer (Beecher Instruments, Sun Prairie, WI, USA). Three targeted core samples with a 1 mm diameter were punched from each specimen and arrayed on a recipient paraffin block, which was then cut into sections (4 µm) and placed on glass slides.
A total of 234 PTs and 639 regional MLN samples from the 234 selected T3N1–3M0 ESCC patients were utilized. A median of two (range 1–17) MLNs were resected from the patients. Each sample of H&E-stained sections was reviewed randomly from a single selected paraffin block to define representative tumor regions. In patients with only one MLN, the right MLN was chosen for TMA construction. However, in patients with multiple MLNs, the appropriate MLN that satisfied the aforementioned criteria was randomly selected. Altogether, 163 pairs of surgically resected ESCC PTs and their corresponding MLNs, as well as 71 PTs without eligible MLNs, were used.
Immunohistochemistry (IHC)
IHC was performed using an IHC kit (Maxim, Fuzhou, People’s Republic of China); the detailed procedure was described in our previous study.29 In brief, after retrieving antigen and non-specific binding blocking, the tissue slides were incubated with rabbit polyclonal anti-HDAC6 (Santa Cruz Biotechnology Inc., Dallas, TX, USA; 1:50 dilution) at 4°C overnight. Then, a biotinylated goat anti-rabbit IgG was used as the secondary antibody, with incubation for 1 hour at 37°C, followed by a horseradish peroxidase conjugate streptavidin–peroxidase working solution for 20 minutes at 37°C. Finally, the slides were reacted with diaminobenzidine (Sigma-Aldrich, St Louis, MO, USA) and counterstained with hematoxylin. Negative controls were prepared using a normal rabbit IgG to replace the primary antibody. Protein expression of HDAC6 was recorded as negative if no staining was present in tumor cells; otherwise, it was recorded as positive.
Two experienced pathologists evaluated the HDAC6 expression in tumor cells independently, blinded to the patient’s clinicopathological information. A semiquantitative system consisting of staining intensity and proportion of positive cells on each slide was used to score the HDAC6 expression. Each score was calculated as “I × Prop”, in which I stands for staining intensity, stratified as: none (0), weak (1), moderate (2), and strong (3), and Prop represents the percentage of positive cells (0–100) in at least 200 cancer cells counted. Therefore, the score was calculated between 0 and 300. The expression level of each case was the average of the scores determined by the two pathologists.
Statistical analyses
Statistical analyses were performed using SPSS version 16.0 (SPSS Inc., Chicago, IL, USA) or MedCalc 9.6.2.0 (MedCalc Software, Mariakerke, Belgium). A matched-pair Wilcoxon test was used to compare HDAC6 expression in paired PTs and MLNs. Receiver operating characteristics (ROC) curves were used to select the optimal cutoff value of HDAC6 expression in PTs and MLNs. The optional cutoff value maximizes both the sensitivity and specificity for survival outcome in the 18 months following the operation. The relationship between HDAC6 expression and clinicopathological characteristics was analyzed by the chi-squared test. Overall survival (OS) was calculated based on the time of surgery to the time of death from any cause, censoring patients who were still alive at the time of the last follow-up (June 4, 2016). Disease-free survival (DFS) was defined as the time from surgery to any regional relapse or distal metastasis, censoring patients who still had an absence of any malignancy at the last follow-up. Survival curves were analyzed by the Kaplan–Meier method and log-rank test. Multivariate analysis was performed using the Cox proportional hazard modes with potential factors whose P-values were less than 0.10 in the univariate analyses, constructed with the forward stepwise method. The result was considered significant when the two-tailed P-value was less than 0.05.
Results
Patients’ characteristics
Owing to the losses of cores during the IHC procedures, nine pairs of PTs and MLNs and 16 PTs without paired MLNs were excluded from the analyses. The remaining TMA included 155 PTs with paired MLNs and 54 PTs without paired MLNs. There were 172 male and 37 female patients, with a median age of 58 years. All of them were node positive, including N1 in 118 patients, N2 in 69 patients, and N3 in 22 patients.
The median length of follow-up for surviving patients was 140 (18–195) months. During the follow-up period, 186 patients (89.0%) died. The 5-year OS and DFS rates were 17.7% and 15.8%, respectively.
HDAC6 expression in ESCC PTs and MLNs
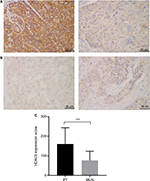
HDAC6 protein was found to be mainly localized to the cytoplasmic region (Figure 1A and B). Positive HDAC6 expression was detected in 96.7% (202/209) and 95.5% (148/155) of ESCC PTs and MLNs, respectively. However, based on their IHC score, HDAC6 expression in MLNs was significantly decreased compared to their paired PTs (Wilcoxon matched-pair signed-rank test, P<0.001) (Figure 1C).
According to HDAC6 expression in ESCC PTs and MLNs, ROC curves were drawn to determine the optimal cutoff value with the best discriminatory power for prediction of survival outcome. As shown in Figure 2, the cutoff score was 156.67 and 76.67 in PTs and MLNs, respectively, with values above these indicating high HDAC6 expression. Using this criterion, high HDAC6 expression was observed in 48.8% (102/209) of the ESCC PTs and 49.0% (76/155) of MLNs. When performing the comparison in 155 paired samples, we found that 50.3% (78/155) of PTs had high HDAC6 expression, but only 53.8% (42/78) of them retained high expression in their corresponding MLNs (P=0.228). HDAC6 expression (high vs low) in ESCC PTs and MLNs was analyzed with regard to their clinicopathological parameters (Table 1). No significant association was obtained between the high and low HDAC6 expression groups for gender, age, smoking, drinking, location, length, grade, and pN stage, in either PTs or MLNs.
HDAC6 expression and ESCC patient survival
Based on Kaplan–Meier data analysis, higher HDAC6 expression in ESCC MLNs was significantly associated with poorer OS (P=0.011) and DFS (P=0.012). The 5-year OS rates for high and low HDAC6 expression in MLNs were 12.1% and 21.4%, respectively (Figure 3A and B). However, neither the OS (P=0.595) nor the DFS (P=0.842) was significantly influenced by the HDAC6 expression level of ESCC PTs. The 5-year OS rates for high and low HDAC6 expression in PTs were 17.4% and 18.0%, respectively (Figure 3C and D).
As shown by the univariate analysis in Table 2, age and pN stage were also significant prognostic factors for OS (age, P=0.010; stage, P=0.006) and DFS (age, P=0.013; stage, P<0.001). Tumor grade was significantly correlated with DFS (P=0.041) but not with OS (P=0.068). Multivariate analysis using the Cox proportional hazards regression model with factors that might affect survival determined by univariate analysis showed that only pN stage and HDAC6 expression in MLNs were independent prognostic factors of OS (pN stage, P=0.004; HDAC6, P=0.029) and DFS (pN stage, P<0.001; HDAC6, P=0.033) in N+ ESCC patients (Table 2).
Subgroup analysis revealed that the significant prognostic value of HDAC6 expression in MLNs was only pronounced in pN1 patients (Figure 4A and B). In patients with pN2 and pN3, there was no significant difference in OS or DFS between high and low HDAC6 expression in MLNs (Figure 4C–F).
Discussion
Although adjuvant therapy following surgery has been validated to benefit N+ ESCC patients, their prognosis is still dismal.30–32 Multiple approaches have been made to improve the outcome for these patients; elucidating the molecular mechanisms for precision medicine is one of these options. However, most of the previous studies only focused on the biological characteristics of the PT. Although gene or protein expression in the PT is often related to lymphatic, distant metastases and prognosis, it has failed to show a similar predictive value in N+ ESCC patients.33,34 Tumor heterogeneity may be part of the reason for this phenomenon. It is now believed that intratumor heterogeneity reflects the ongoing linear and branching evolution, resulting in multiple simultaneous subclones that may individually be capable of giving rise to metastasis.35 In this context, somatic genetic alterations are restricted or enriched in the metastatic lesions compared to their respective PTs.36,37 Our previous study also demonstrated the different epithelial–mesenchymal phenotypes between PTs and their corresponding MLNs. The transition of tumor cells from mesenchymal to epithelial phenotypes may be a key factor in the formation of metastasis.29 In the present study, we again confirmed that there are differences in molecular expression in ESCC PTs and corresponding MLNs. The HDAC6 expression in MLNs was significantly decreased.
In addition, we found that HDAC6 expression in MLNs, but not in PTs, was associated with both the DFS and OS in survival analyses. This suggests that more attention should be paid to genomic expression in MLN in N+ patients, not only to elucidate the mechanism of cancer cell migration but also to help determine patient prognosis and guide treatment. Nonetheless, only a few studies have focused on this issue. In non-small-cell lung cancer with lymph-node metastases, Kilvaer et al38 found that a high level of intraepithelial CD45RO+ tumor infiltrative lymphocytes in MLNs was an independent positive prognostic factor for disease-specific patient survival. In stage II/III lymph-node-positive breast cancer patients, Bonin et al39 determined that keratin 8 expression in MLNs, but not in PTs, indicated better survival. Our previous study also revealed that high expression of C-terminal Hsp-interacting protein (CHIP) in MLNs suggests a poor prognosis for patients with resected T3N1–3M0 ESCC.40 Taking our findings together with these previous studies, we strongly recommend examining the genomic profiling of MLNs when assessing prognosis in lymph-node-positive cancer patients.
HDAC6 has been shown to be upregulated in a diverse number of tumors and cancer cell lines, suggesting an important role for this enzyme in cancer. It is essential in maintaining oncogenic phenotype and promoting anchorage-independent proliferation in transformed cells41 and leads to increased cell motility.42 In an ESCC in vitro model, Li et al28 confirmed its role in tumor progression by showing that cell proliferation and migration could both be significantly reduced after HDAC6 inhibition. Tao et al43 found that HDAC6 facilitated ESCC development by regulating the acetylation of HSP90, and coadministration of HSP90 and HDAC6 inhibitors strongly inhibited tumor growth in mice. HDAC6 inhibitors, such as ricolinostat and ACY-241, stand apart from broad-spectrum HDAC inhibitors because of their druggability and unique function with the cells. Unlike other pan-HDAC inhibitors with adverse effects including hematological toxicity and QT prolongation, highly selective HDAC6 inhibitors are considered to have more potential for clinical use.44 A number of clinical trials utilizing selective HDAC6 inhibitors are underway for treating multiple myeloma and lymphoid malignancies. Therefore, HDAC6 is not only a biomarker predicting patients’ outcome but also a potential therapeutic target.
Several limitations exist in this study. First, in patients with more than one MLN, samples were collected randomly. The heterogeneity between the different MLNs may impart some bias to our results. Second, most of the participants were from southern China, which may limit the generalization of our findings to other populations. Finally, the small sample size and retrospective nature of our study suggest the need to perform a large-scale prospective study to confirm our results.
Conclusion
Although HDAC6 was found to be highly expressed in most PTs and MLNs in ESCC, a substantial discordance between them was still present. HDAC6 expression was decreased in MLNs compared to their paired PTs and may serve as an independent predictor for prognosis of complete surgically resected T3N1–3M0 ESCC patients. More attention should be paid to HDAC6 expression in metastatic tumors for prognostic prediction and to the potential for HDAC6 inhibitor therapy in ESCC patients.
Abbreviations
N+, node involvement; MLN, metastatic lymph node; ESCC, esophageal squamous cell cancer; HDAC, histone deacetylase; PT, primary tumor; TMA, tissue microarray; IHC, immunohistochemistry; ROC, receiver operating characteristics; OS, overall survival; DFS, disease-free survival.
Acknowledgments
This work was supported by the National Science Foundation of China (grant nos 81672356 and 81602105), Guangzhou Science Technology and Innovation Commission (grant no. 201610010127), Guangdong Talents Special Support Program (grant no. 201629038), the Fundamental Research Funds for the Central Universities (grand nos 15ykpy34 and 16ykjc39), and Guangdong Esophageal Cancer Institute Science and Technology Program (grant no. M201701).
Author contributions
XX and KJL constructed the tissue microarrays, designed the study, analyzed the data, and drafted the manuscript. YL carried out data acquisition and performed the statistical analysis. YHL evaluated the protein expression and calculated the score of each sample. SSZ and XYX helped to construct the tissue microarrays and performed the immunohistochemistry. JW conceived the study, participated in its design and coordination, and helped to draft the manuscript. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. | ||
Rice TW, Ishwaran H, Ferguson MK, Blackstone EH, Goldstraw P. Cancer of the Esophagus and Esophagogastric Junction: An Eighth Edition Staging Primer. J Thorac Oncol. 2017;12(1):36–42. | ||
Sjoquist KM, Burmeister BH, Smithers BM, et al; Australasian Gastro-Intestinal Trials Group. Survival after neoadjuvant chemotherapy or chemoradiotherapy for resectable oesophageal carcinoma: an updated meta-analysis. Lancet Oncol. 2011;12(7):681–692. | ||
Sugawara K, Yamashita H, Uemura Y, et al. Numeric pathologic lymph node classification shows prognostic superiority to topographic pN classification in esophageal squamous cell carcinoma. Surgery. 2017;162(4):846–856. | ||
Peng J, Wang WP, Dong T, et al. Refining the Nodal Staging for Esophageal Squamous Cell Carcinoma Based on Lymph Node Stations. Ann Thorac Surg. 2016;101(1):280–286. | ||
Fu X, Liu Q, Luo K, et al. Lymph node station ratio: Revised nodal category for resected esophageal squamous cell carcinoma patients. J Surg Oncol. 2017;116(7):939–946. | ||
Wang F, Zheng Y, Wang Z, et al. Nodal Skip Metastasis in Esophageal Squamous Cell Carcinoma Patients Undergoing Three-Field Lymphadenectomy. Ann Thorac Surg. 2017;104(4):1187–1193. | ||
Rice TW, Ishwaran H, Hofstetter WL, et al. Esophageal Cancer: Associations With (pN+) Lymph Node Metastases. Ann Surg. 2017;265(1):122–129. | ||
Wood SL, Pernemalm M, Crosbie PA, Whetton AD. Molecular histology of lung cancer: from targets to treatments. Cancer Treat Rev. 2015;41(4):361–375. | ||
Ades F, Zardavas D, Bozovic-Spasojevic I, et al. Luminal B breast cancer: molecular characterization, clinical management, and future perspectives. J Clin Oncol. 2014;32(25):2794–2803. | ||
Ciombor KK, Wu C, Goldberg RM. Recent therapeutic advances in the treatment of colorectal cancer. Annu Rev Med. 2015;66(1):83–95. | ||
Su H, Hu N, Yang HH, et al. Global gene expression profiling and validation in esophageal squamous cell carcinoma and its association with clinical phenotypes. Clin Cancer Res. 2011;17(9):2955–2966. | ||
Delcuve GP, Khan DH, Davie JR. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenetics. 2012;4(1):5. | ||
Zou H, Wu Y, Navre M, Sang BC. Characterization of the two catalytic domains in histone deacetylase 6. Biochem Biophys Res Commun. 2006;341(1):45–50. | ||
Zhang Y, Gilquin B, Khochbin S, Matthias P. Two catalytic domains are required for protein deacetylation. J Biol Chem. 2006;281(5):2401–2404. | ||
Bali P, Pranpat M, Bradner J, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280(29):26729–26734. | ||
Zhang X, Yuan Z, Zhang Y, et al. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell. 2007;27(2):197–213. | ||
Parmigiani RB, Xu WS, Venta-Perez G, et al. HDAC6 is a specific deacetylase of peroxiredoxins and is involved in redox regulation. Proc Natl Acad Sci U S A. 2008;105(28):9633–9638. | ||
Nogués L, Reglero C, Rivas V, et al. G Protein-coupled Receptor Kinase 2 (GRK2) Promotes Breast Tumorigenesis Through a HDAC6-Pin1 Axis. EBioMedicine. 2016;13:132–145. | ||
Asthana J, Kapoor S, Mohan R, Panda D. Inhibition of HDAC6 deacetylase activity increases its binding with microtubules and suppresses microtubule dynamic instability in MCF-7 cells. J Biol Chem. 2013;288(31):22516–22526. | ||
Wang XX, Wan RZ, Liu ZP. Recent advances in the discovery of potent and selective HDAC6 inhibitors. Eur J Med Chem. 2018;143:1406–1418. | ||
Jung KH, Noh JH, Kim JK, et al. Histone deacetylase 6 functions as a tumor suppressor by activating c-Jun NH2-terminal kinase-mediated beclin 1-dependent autophagic cell death in liver cancer. Hepatology. 2012;56(2):644–657. | ||
Li D, Sun X, Zhang L, et al. Histone deacetylase 6 and cytoplasmic linker protein 170 function together to regulate the motility of pancreatic cancer cells. Protein Cell. 2014;5(3):214–223. | ||
Lin TW, Chen MT, Lin LT, et al. TDP-43/HDAC6 axis promoted tumor progression and regulated nutrient deprivation-induced autophagy in glioblastoma. Oncotarget. 2017;8(34):56612–56625. | ||
Seo J, Min SK, Park HR, et al. Expression of Histone Deacetylases HDAC1, HDAC2, HDAC3, and HDAC6 in Invasive Ductal Carcinomas of the Breast. J Breast Cancer. 2014;17(4):323–331. | ||
Yang CJ, Liu YP, Dai HY, et al. Nuclear HDAC6 inhibits invasion by suppressing NF-κB/MMP2 and is inversely correlated with metastasis of non-small cell lung cancer. Oncotarget. 2015;6(30):30263–30276. | ||
He Q, Li G, Wang X, et al. A Decrease of Histone Deacetylase 6 Expression Caused by Helicobacter Pylori Infection is Associated with Oncogenic Transformation in Gastric Cancer. Cell Physiol Biochem. 2017;42(4):1326–1335. | ||
Li N, Tie XJ, Liu PJ, et al. Effects of down-regulation of HDAC6 expression on proliferation, cell cycling and migration of esophageal squamous cell carcinoma cells and related molecular mechanisms. Asian Pac J Cancer Prev. 2013;14(2):685–689. | ||
Wen J, Luo KJ, Liu QW, et al. The epithelial-mesenchymal transition phenotype of metastatic lymph nodes impacts the prognosis of esophageal squamous cell carcinoma patients. Oncotarget. 2016;7(25):37581–37588. | ||
Zhang SS, Yang H, Xie X, et al. Adjuvant chemotherapy versus surgery alone for esophageal squamous cell carcinoma: a meta-analysis of randomized controlled trials and nonrandomized studies. Dis Esophagus. 2014;27(6):574–584. | ||
Lyu X, Huang J, Mao Y, et al. Adjuvant chemotherapy after esophagectomy: is there a role in the treatment of the lymph node positive thoracic esophageal squamous cell carcinoma? J Surg Oncol. 2014;110(7):864–868. | ||
Qin RQ, Wen YS, Wang WP, et al. The role of postoperative adjuvant chemotherapy for lymph node-positive esophageal squamous cell carcinoma: a propensity score matching analysis. Med Oncol. 2016;33(4):31. | ||
Xie X, Zhang SS, Wen J, et al. Prognostic value of HOXB7 mRNA expression in human oesophageal squamous cell cancer. Biomarkers. 2013;18(4):297–303. | ||
Chen YF, Xie JD, Jiang YC, et al. The Prognostic Value of Peripheral Benzodiazepine Receptor in Patients with Esophageal Squamous Cell Carcinoma. J Cancer. 2017;8(16):3343–3355. | ||
Podlaha O, Riester M, De S, Michor F. Evolution of the cancer genome. Trends Genet. 2012;28(4):155–163. | ||
Schrijver W, Selenica P, Lee JY, et al. Mutation profiling of key cancer genes in primary breast cancers and their distant metastases. Cancer Res. 2018;78(12):3112–3121. | ||
Saber A, Hiltermann TJN, Kok K, et al. Mutation patterns in small cell and non-small cell lung cancer patients suggest a different level of heterogeneity between primary and metastatic tumors. Carcinogenesis. 2017;38(2):144–151. | ||
Kilvaer TK, Paulsen EE, Khanehkenari MR, et al. The presence of intraepithelial CD45RO+ cells in resected lymph nodes with metastases from NSCLC patients is an independent predictor of disease-specific survival. Br J Cancer. 2016;114(10):1145–1151. | ||
Bonin S, Pracella D, Barbazza R, Sulfaro S, Stanta G. In stage II/III lymph node-positive breast cancer patients less than 55 years of age, keratin 8 expression in lymph node metastases but not in the primary tumour is an indicator of better survival. Virchows Arch. 2015;466(5):571–580. | ||
Wen J, Luo KJ, Hu Y, Yang H, Fu JH. Metastatic lymph node CHIP expression is a potential prognostic marker for resected esophageal squamous cell carcinoma patients. Ann Surg Oncol. 2013;20(5):1668–1675. | ||
Aldana-Masangkay GI, Sakamoto KM. The role of HDAC6 in cancer. J Biomed Biotechnol. 2011;2011:875824. | ||
Saji S, Kawakami M, Hayashi S, et al. Significance of HDAC6 regulation via estrogen signaling for cell motility and prognosis in estrogen receptor-positive breast cancer. Oncogene. 2005;24(28):4531–4539. | ||
Tao H, Chen YY, Sun ZW, Chen HL, Chen M. Silence of HDAC6 suppressed esophageal squamous cell carcinoma proliferation and migration by disrupting chaperone function of HSP90. J Cell Biochem. 2018;119(8):6623–6632. | ||
Batchu SN, Brijmohan AS, Advani A. The therapeutic hope for HDAC6 inhibitors in malignancy and chronic disease. Clin Sci (Lond). 2016; 130(12):987–1003. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.