Back to Journals » OncoTargets and Therapy » Volume 13
Highly Sensitive Droplet Digital PCR Method for Detection of de novo EGFR T790M Mutation in Patients with Non-Small Cell Lung Cancer
Authors Wang X , Li X, Guo H , Zhu L, Peng Z, Wang J, Yang F, Guo Y
Received 14 June 2020
Accepted for publication 23 September 2020
Published 22 October 2020 Volume 2020:13 Pages 10621—10630
DOI https://doi.org/10.2147/OTT.S267677
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Carlos E Vigil
Xun Wang,1,* Xiao Li,1,* Haifa Guo,1,* Lingxiang Zhu,2 Zhiyong Peng,3 Jun Wang,1 Fan Yang,1 Yong Guo4
1Department of Thoracic Surgery, Peking University People’s Hospital, Peking University, Beijing 100044, People’s Republic of China; 2National Research Institute for Health and Family Planning, Beijing 100081, People’s Republic of China; 3TargetingOne Corporation, Beijing 100190, People’s Republic of China; 4Department of Biomedical Engineering, School of Medicine, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Tsinghua University, Beijing 100084, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Fan Yang
Department of Thoracic Surgery, Peking University People’s Hospital, Peking University, Beijing 100044, People’s Republic of China
Email [email protected]
Yong Guo
Department of Biomedical Engineering, School of Medicine, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Tsinghua University, Beijing 100084, People’s Republic of China
Email [email protected]
Objective: The aim of this study was to investigate the allelic relation between de novo T790M and concomitant sensitizing EGFR mutations in EGFR-TKIs naïve NSCLCs and to explore whether the formalin-fixed and paraffin-embedded (FFPE) materials affect the detection of de novo T790M mutation.
Methods: Specimens of 300 consecutive EGFR-TKI naïve NSCLCs who received surgical resection between January 2016 and June 2018 were retrospectively investigated. All the snap-frozen tumor tissues from 300 NSCLCs were screened by droplet digital PCR (ddPCR) for the detection of de novo T790M mutation. The allelic relation between de novo T790M mutation and concomitant sensitizing EGFR mutations was also investigated. Furthermore, we assessed de novo T790M mutation in paired FFPE specimens of 50 patients which included tumor tissues and paired normal lung tissues of the pretreatment NSCLCs to investigate whether FFPE materials affect the detection of de novo T790M mutation.
Results: The de novo T790M mutation was observed in four patients which included one patient of single de novo T790M mutation and three patients of de novo T790M mutation coexisting with L858R mutation. The incidence of de novo T790M in pretreatment NSCLCs who harboring EGFR mutations was 2.9% (4/139). All the de novo T790M mutations were detected in cis with the concomitant L858R mutations for the three NSCLCs. Our ddPCR method demonstrated that the frequency of de novo T790M mutation was ranging from 0.1% to 0.5% among 90% (45/50) of the FFPE tumor samples and 92% (46/50) of the paired FFPE adjacent normal lung samples. The frequency of de novo T790M mutation in the paired snap-frozen samples was all below 0.1%.
Conclusion: Our study demonstrated that most de novo T790M mutations were detected in cis with concomitant sensitizing mutations for pretreatment NSCLCs. Analytical cut-off of ddPCR assay for FFPE specimens should be validated carefully considering the possibility of FFPE-derived artificial gene mutations.
Keywords: non-small cell lung cancer, NSCLC, droplet digital PCR, de novo T790M, acquired T790M, EGFR mutation
Introduction
Sensitizing mutations in the epidermal growth factor receptor (EGFR) gene are the most frequent oncogenic alterations in non-small cell lung cancer (NSCLC) presenting in approximately 10%-15% of Caucasians and 40%-50% of Asian patients with lung adenocarcinoma.1–4 The most common sensitizing mutations in EGFR are in-frame deletions around the amino acids Leu Arg Glu Ala in exon 19 (19Del) and the L858R point-mutation in exon 21 (L858R), which are considered as classical EGFR mutations, accounting for approximately 85% of sensitizing EGFR mutations.5 NSCLC patients with sensitizing EGFR mutations demonstrate improved objective responses and prolonged progression-free survival (PFS) under the treatment with EGFR tyrosine kinase inhibitors (EGFR-TKIs), such as gefitinib, afatinib, and osimertinib.6–8
The uncommon EGFR mutations (such as G719X, L861Q, S768I, and T790M) comprise approximately 10% - 18% of all EGFR mutations.9,10 The emergence of T790M in exon 20 of EGFR has been demonstrated to be associated with acquired EGFR-TKI resistance which accounts for more than 50% of such cases.11 Low frequency de novo T790M mutation has been detected in some EGFR-TKI treatment naïve NSCLC patients using several different highly sensitive methods.12,13 However, there were discrepant results for the incidence of de novo T790M in different literatures with a wide range from 1% to 79%.12,13 The therapeutic efficacy of the third-generation EGFR-TKIs, such as osimertinib, is better than the earlier-generation EGFR-TKIs for the patients with concomitant de novo T790M and sensitizing EGFR mutations.14 There is still controversy on whether the de novo T790M occurs occasionally or coexists with the presence of other sensitizing EGFR mutations. It is important to identify the allelic relation between de novo T790M and concomitant sensitizing EGFR mutations among clinical specimens for the purpose to better understand the intrinsic mechanism of earlier-generation EGFR-TKIs resistance. However, literatures regarding this issue are limited and deserve further investigation. Moreover, it is still not clear whether the de novo T790M detection rate could be affected by formalin-fixed and paraffin-embedded (FFPE) samples which may cause artificial gene mutations (C-T or G-A transitions).15 Therefore, it is necessary to develop a reliable and stable platform with high sensitivity for the detection of de novo T790M in EGFR-TKI treatment naïve NSCLC patients.
Droplet digital PCR (ddPCR) is a new generation of PCR technique with high sensitivity and specificity for the detection of EGFR mutations.15,16 In the present study, clinical specimens of 300 consecutive Chinese EGFR-TKI treatment naïve NSCLC patients were retrospectively collected. We investigated the prevalence and concomitant sensitizing EGFR mutations of de novo T790M mutation among them using a highly sensitive and specific ddPCR assay. Moreover, we investigated the allelic relation between de novo T790M and concomitant sensitizing EGFR mutations among the clinical specimens of EGFR-TKIs naïve NSCLC patients and explored whether the FFPE materials affect the detection of de novo T790M mutation.
Materials and Methods
Patients and Specimen
Specimens of 300 consecutive Chinese EGFR-TKI treatment naïve NSCLC patients who received surgical resection at Peking University People’s Hospital between January 2016 and June 2018 were retrospectively analyzed in this study. For the first 250 patients (Group A), only snap-frozen tumor tissues (stored at −80 °C) were analyzed. In the next 50 patients (Group B), both FFPE tumor tissues and snap-frozen tumor tissues (stored at −80 °C) were analyzed, and two types of the adjacent normal lung tissues were also analyzed simultaneously (Figure 1). All patients provided a written informed consent. The study was approved by the Ethics Committee of Peking University People’s Hospital (Approved number: 2019 PHB 259–01) and was conducted in accordance with the Declaration of Helsinki.
 |
Figure 1 The flowchart of the study design. |
ddPCR Analysis
The ddPCR platform system (TargetingOne® Biotech. Co. Ltd. Beijing, China) which utilized for this evaluation includes the Drop Maker and the Chip Reader. The Drop Maker is equipped with a microfluidic chip to quickly prepare the aqueous phase sample into a nanoliter drops. The number of droplets is related to the sample volume, and about 50,000–60,000 droplets can be prepared from 30 microliters of aqueous phase sample. The Chip Reader uses the principle of laser confocal scanning and is equipped with a microfluidic chip to locate and identify nanoliter droplets accurately and quickly and obtain their fluorescence signal values. After Poisson statistical analysis, the absolute number of positive and negative droplets is provided to derive the exact concentration of the starting target nucleic acid molecule. The Chip Reader is compatible with TaqMan Hydrolysis Probes and EvaGreen Detection and has two channels which include the FAM (6-carboxyfluorescein) and the VIC channels.
The human EGFR gene mutation detection kit can detect 42 common mutations in the 18–21 exons of the EGFR gene. There are six EGFR mutation assays in this kit, including 19Del, L858R, T790M, G719X, L861Q, and S768I assays. The detection sensitivity was 0.1% - 0.5% mutation rate, depending on the type of sample and the type of assay. The reaction components were prepared as follows: 7.5 μL of PCR reagent A (SuperMix), 7.5 μL of PCR reagent B (primers and probes), and 20–50 ng of DNA extracted from FFPE tissues using QIAamp® DNA FFPE Tissue kit (Qiagen, Hilden, Germany) or from frozen tissues using TIANamp® Genomic DNA Kit (TIANGEN Biotech, Beijing, China). Sterilized water was used to make a volume of up to 30 μL. Next, 30 μL of digital PCR reaction mixture and 180 μL of droplet generation oil were added into a droplet generation chip, and the droplet generation was performed using a Drop Maker following the manufacturer’s instructions.
About 100 μL of the resulting droplet emulsion were automatically transferred into an 8-strip PCR tube and amplified in a PTC-200 Thermal Cycler (Bio-Rad, CA). The PCR conditions were as follows: pre-denaturation at 95 °C for 10 minutes, amplification for 40 cycles, with denaturation at 95 °C for 30 seconds, annealing at 60 °C for 1 minute, ending at 12 °C for 10 minutes. After PCR, the 8-strip PCR tube containing the droplets was connected to a droplet detection chip, then the fluorescence signal of droplet was detected on Chip Reader. Finally, the data were subjected to Poisson distribution analysis using the ddPCR software to obtain the target DNA copy number in the samples.
Statistical Analysis
The statistical analysis was performed with Statistical Product and Service Solutions (SPSS) software versions 20.0 (2011; IBM, Armonk, NY, USA). Differences were compared using a t test for continuous clinicopathological variables, and Chi-square test or Fisher’s exact test for categorical mutations and clinicopathological variables. P value < 0.05 was considered statistically significant.
Results
Patient Characteristics
The median age of the cohort was 62 years old (ranging from 35 to 88 years old), which including 171 males and 129 females. Seventy-three (24.3%) were smokers, and more than 50% patients were stage I NSCLC. There were 224 (74.7%) adenocarcinomas, 72 (24.0%) squamous cell carcinomas, and 4 (1.3%) adenosquamous carcinomas. EGFR mutations were mainly distributed in non-smoker, female patients, and adenocarcinomas. The baseline characteristics of the 300 patients are listed in Table 1.
 |
Table 1 Baseline Clinical Characteristics of the NSCLC Patients (N = 300) |
The Condition of EGFR Mutations in Snap-Frozen Tumor Tissue
Snap-frozen tumor tissues from 300 EGFR-TKI naïve NSCLC patients were screened by ddPCR for the detection of EGFR mutations including 19Del, L858R, T790M, G719A, S768I, and L861Q. EGFR mutations were observed in 139 of 300 (46.3%) patients. Single EGFR mutations were detected in 134 patients, including 49 (16.3%) patients with 19Del, 79 (26.3%) patients with L858R, 4 (1.3%) patients with L861Q, 1 (0.3%) with T790M, and 1 (0.3%) with G719A. Concurrent EGFR mutations were detected in 5 (2.3%) patients including three patients with EGFR L858R+T790M mutations, one patient with EGFR G719A+L861Q mutations, and one patient with EGFR G719A+S768I mutations (Table 2). All the uncommon EGFR mutations were detected in patients with adenocarcinoma. Patients characteristics and pathologic stages are listed in Table 3.
 |
Table 2 A Comprehensive View of the EGFR Mutations |
 |
Table 3 The Characteristics of the Patients with Uncommon EGFR and de novo T790M Mutations |
De novo GFR T790M mutation was observed in 4 (4/300, 1.3%) patients with an incidence of 2.9% (4/139) in the EGFR-mutated cohort, including one patient with single de novo T790M mutation and three patients harboring de novo T790M mutation coexisting with L858R mutation which suggested that the de novo T790M was more likely to coexist with EGFR L858R mutation (Table 3).
Evaluation of the Allelic Relation Between de novo T790M and Sensitizing EGFR Mutations in NSCLC Specimens
Furthermore, we investigated the allelic relation between de novo T790M and concomitant sensitizing EGFR mutations. In our study, concomitant occurrence of de novo T790M and L858R was detected in three individuals and no patient was found to have a de novo T790M combined with 19Del. Total RNA was obtained from the three patients with de novo T790M combined with L858R. A reverse transcription (RT) ddPCR-based method was designed to evaluate the allelic relation between the two EGFR mutations. Analysis was performed with the mutation-specific probes which were designed as upstream primers of the T790M and the probe targeting de novo T790M of EGFR (labeled with FAM) combined with downstream primers of the L858R and the probe targeting L858R of EGFR (labeled with VIC). Double positive signals for the de novo T790M and L858R of EGFR were detected among the three NSCLC patients which demonstrated that all the de novo T790M mutations were present together with the EGFR L858R mutations on the same allele (Figure 2).
Influence of FFPE Materials on the Detection of de novo T790M Mutation
For the 50 patients in group B, FFPE sample of tumors and their adjacent normal lung tissues were also analyzed. Droplet digital PCR was used for the detection of EGFR mutations including19Del, L858R, and T790M. Together with the detection results of snap-frozen samples from the same cohort, EGFR mutations were detected in 26 NSCLC tumor samples regardless of the specimen type. And most patients (24/26) with common sensitizing EGFR mutations had concordant mutations between the snap-frozen tumor samples and FFPE tumor tissue samples. Surprisingly, de novo T790M mutation was detected in 90% (45/50) of the FFPE tumor samples and 92% (46/50) of the paired FFPE adjacent normal lung samples with a frequency of 0.1%-0.5% using our ddPCR method (Figure 3). However, the frequency of de novo T790M mutation in the paired snap-frozen samples was all below 0.1% which were identified as de novo T790M-negative according to the identifying standard for frozen samples.
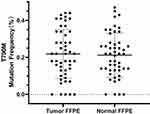 |
Figure 3 The de novo T790M mutant abundance in the FFPE tumor tissues samples and FFPE adjacent normal lung samples. |
Discussion
The emergence of EGFR T790M mutation is a major cause of acquired resistance to first-generation EGFR-TKIs for NSCLC patients harboring sensitizing EGFR mutations. Moreover, several studies suggested that the de novo T790M mutation was present in a minor subgroup among EGFR-TKI treatment naïve NSCLC patients.17,18 It is reported that the incidence of de novo T790M mutation could be up to about 79% among sensitizing mutation-positive NSCLC patients before EGFR-TKI treatment using highly sensitive methods, such as ddPCR or colony hybridization, and most of the de novo T790M mutations were below 0.1% in their frequency.13,17 But studies from other groups reported that they detected de novo T790M mutations in 0.5–1% EGFR-TKI naïve NSCLC patients by ARMS or targeted sequencing with sensitivity of about 1%.12,14 The great difference of reported incidence of de novo T790M mutation may come from the detecting sensitivity of the methods. This hypothesis was supported by the study from Inukai et al in which only one T790M mutant case was detected by the direct sequencing, while 9 additional cases among 280 cases were detected using mutant-enriched PCR (ME-PCR, sensitivity of 0.1%).18 However, the accurate incidence of de novo T790M is still largely unknown.12,13,19 Further, the influence of FFPE sample on the detection rate of T790M, as well as the allelic relationship of de novo T790M and concurrent sensitizing mutations have not been well proved. In this study, snap-frozen tumor tissue from 300 consecutive Chinese EGFR-TKI naïve NSCLC patients were tested using ddPCR to investigate the prevalence of de novo T790M mutation. The incidence of de novo T790M in pretreatment NSCLC patients who harboring EGFR mutations was 2.9% (4/139) with adenocarcinoma in our cohort, which was consistent with Ye’s report using ME-PCR with sensitivity of 0.1%.
For the purpose to detect the accompanying mutations coexisted with de novo T790M mutation, two common sensitizing EGFR mutations (L858R and 19Del) and three most frequent uncommon EGFR mutations (G719A, S768I, and L861Q) were detected simultaneously. Consistent with previous studies,12,14,20 we found that most de novo T790M mutation coexist with L858R mutation (75%, 3/4). De novo T790M mutation could also co-exist with 19 Del, as well as other uncommon EGFR mutations.12,20 The reason we did not find co-existing de novo T790M and 19 Del in our study perhaps because of the small sample size and rare de novo T790M cases. However, it was reported that acquired T790M mutation always coexist with 19Del.20–22 Tian’s study22 reported that the ratio of allele frequency (Relative allele frequency, RAF) of the T790M mutation to the EGFR sensitizing mutation was different between the de novo and acquired T790M mutations (86.1% vs 22.3%, P< 0.0001). Hata et al23 reported that the T790M mutations could occur both by the selection of pre-exist T790M clones and the genetic evolution from drug-tolerant cells, and the two types of T790M positive cells developed at different times and showed differential response to the EGFR inhibition which suggested that they might have different molecular characteristics. The intrinsic mechanism of the different trends for the association between T790M mutations (de novo or acquired) and sensitizing EGFR mutations remains unclear.
The only study to investigate the allelic relation between de novo T790M and the concurrent sensitizing EGFR mutations in the treatment naïve NSCLC patients to date was reported by Hidakaet al,24 and only one such case was reported in their study. In our study, we investigated the allelic relation between de novo T790M and concurrent EGFR L858R mutations among the three clinical specimens from EGFR-TKI naïve NSCLC in our cohort. Our result further conformed that all de novo T790M mutations were present on the same EGFR allele with the sensitizing EGFR mutations in the pretreatment NSCLC patients. Theoretically, the in cis compound mutations of de novo T790M and L858R mutations might affect the binding of first- or second- generation EGFR-TKIs to the ATP-binding site of EGFR. For the NSCLC patients with de novo T790M and sensitizing EGFR mutation, the use of earlier-generation EGFR-TKIs would lead to a decrease of sensitive target cells and an enrichment of resistant double-mutated tumor cells.4,24
Watanabe et al17 reported that they detected de novo T790M mutations in 79.9% (298/373) FFPE tumor specimens of pretreatment NSCLC patients, using ddPCR with a sensitivity of 0.001%. In Iwama’s study,25 the prevalence of de novo T790M mutation was 100% (25/25) among the pretreatment FFPE tissue specimens by ddPCR technique with a sensitivity of 0.01%. Such a prevalence of de novo T790M mutation might be caused by highly sensitive methods in clinical molecular testing. However, the influence of FFPE sample on the detection of EGFR T790M mutation was another concern when evaluating the prevalence of de novo T790M. It was reported that the formalin fixation could led to false-positive artificial mutations, and most of them were C-T or G-A transitions mutations which could happen in the EGFR T790M mutation.15 Inukai et al18 identified 4.2% (4/95) of de novo T790M mutation rate using mutant-enriched PCR method (ME-PCR, 0.1% sensitivity) and in this study, approximately 80% of samples were snap-frozen specimens. However, in Ye’s study,15 the prevalence of de novo T790M mutation was 41.7% (15/36) in FFPE specimens using the same ME-PCR assay with a sensitivity of 0.1%. Moreover, Ye et al demonstrated that the incidence of de novo T790M mutation in FFPE specimens was much higher than that in paired frozen samples, and the artificial T790M mutation could be detected in 48.8% (16/33) FFPE adjacent normal samples of EGFR-TKI naïve NSCLC patients.
However, there was no subsequent studies to provide more evidence to clarify this issue. In the present study, we assessed the de novo T790M in 50 pairs of frozen and FFPE tumor and adjacent normal tissues specimens of EGFR-TKI naïve NSCLC patients, using ddPCR with sensitivity of 0.1%. Our ddPCR method demonstrated that the frequency of de novo T790M mutation was ranging from 0.1% to 0.5% among more than 90% of the FFPE tumor and normal lung samples. Meanwhile, the frequency of de novo T790M mutation were all below 0.1% in the paired snap-frozen samples, which was consistent with Ye’s report. Therefore, the artificial mutations caused by the formalin fixation might affect the detection of de novo T790M mutation, and analytical cut-off of ddPCR assay for FFPE specimens should be validated carefully before their application on clinical testing considering the possibility of FFPE-derived artificial gene mutations.
Many studies have demonstrated unfavorable outcome in patients with co-existence of de novo T790M mutation who were treated with first-generation TKIs.14,22,26 However, a previous meta-analysis by Liu et al27 indicated that the de novo T790M mutation did not affect the PFS or the OS in NSCLC patients who harbored sensitizing EGFR mutations and were treated with EGFR-TKIs. An in vitro experiment28 reported that the cell lines of NSCLC harboring T790M and L858R mutations exhibited increased phosphorylated EGFR protein expression compared to single L858R mutation which was linked to the resistance to first- and second-generation EGFR inhibitors. It is presumed that the sensitivity to the earlier-generation EGFR-TKIs would be affected by the abundance of de novo T790M mutation among the NSCLC patients. Therefore, the negative results of the Liu’s meta-analysis might come from the results of some studies with ultra-low allele frequency of de novo T790M. Thus, it is necessary to use appropriate methods, select optimal cut-off values, and confirm positive results by an alternative method in clinical molecular diagnosis.
Some previous studies12,27 reported that patients with de novo T790M mutation could benefit from osimertinib as the first-line treatment. The FLAURA study29 suggested that the median PFS for the sensitizing EGFR mutation positive NSCLCs could be 18.9 months by the initial using of osimertinib. It seemed that to use osimertinib as first-line treatment for sensitizing EGFR mutation positive NSCLCs did not significantly improve the PFS than that of sequential use of earlier-generation EGFR-TKIs followed by third-generation EGFR-TKIs. The planned final analysis of the FLAURA trail30 showed that to use osimertinib as first-line treatment for the sensitizing EGFR mutation positive NSCLCs could have a better median OS than that of first-generation EGFR-TKIs (38.6 vs 31.8 months, P = 0.046). The clinical benefit might be attributed to the existence of de novo T790M mutation and the central nervous system metastases, and various different potential resistance mechanisms which involved drug pressure or genetic evolution.23,30 However, OS benefit with osimertinib was not observed in the subgroup of Asian patients and patients with EGFR L858R mutation.30 Further research is needed to confirm the therapeutic effect of osimertinib to treat late-stage NSCLC with sensitizing EGFR mutation.
Nowadays, the detection of the EGFR T790M mutation is of importance. Firstly, the EGFR-TKI treatment strategy would be affected by the status of the EGFR T790M mutation which is a known mechanism of earlier-generation EGFR-TKIs resistance. Secondly, At the development of resistance to osimertinib, the allelic relation between the EGFR T790M mutation and the EGFR C797S mutation should be identified to determine the subsequent treatment strategy.31 Thirdly, the dynamic monitoring of EGFR T790M mutation during treatment with EGFR-TKI could determine treatment effects.31,32
There are some limitations to this study. Firstly, the selection bias could not be avoided considering the retrospective nature of the study. Secondly, the number of patients with sensitizing EGFR mutations were relatively small and only 3 cases were detected with concurrent de novo T790M and sensitizing EGFR mutations. Thirdly, most of our patients were early-stage NSCLCs and lacking treatment results with TKIs. Fourthly, the spatial heterogeneity of the tumor might have an influence on the detection of the de novo T790M mutation.
Conclusions
In conclusion, we tested the prevalence of de novo T790M mutation in NSCLC using sensitive ddPCR method and snap-frozen tumor tissue, which would offer relative accurate results. We reported the allelic relationship of de novo T790M and concurrent sensitizing mutations in three cases, which was the largest cohort to date with great importance. And our study also evaluated the influence of FFPE specimen on the detection rate of T790M, which could be considered in clinical molecular diagnosis.
Availability of Data and Materials
Fan Yang (corresponding author) can provide all of datasets analyzed during the study on reasonable request. Data Sharing Statement: The datasets will be available from Fan Yang (corresponding author).
Abbreviations
NSCLC, Non-small cell lung cancer; ddPCR, Droplet digital PCR; EGFR, Epidermal growth factor receptor; EGFR-TKIs, Epidermal growth factor receptor tyrosine kinase inhibitors; FFPE, Formalin-fixed and paraffin-embedded; PFS, Progression-free survival; OS, Overall survival.
Ethics Approval and Consent to Participate
This study was approved by the Ethics Committee of Peking University People’s Hospital (Approved number: 2019 PHB 259-01).
Consent for Publication
All authors have approved the submitted manuscript.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work. These authors contributed equally to this work and should be considered co-first authors: Xun Wang, Xiao Li, and Haifa Guo. Co-corresponding authors: Fan Yang and Yong Guo.
Funding
There is no funding to report.
Disclosure
Zhiyong Peng is an employee in TargetingOne Corporation. The authors declare that they have no other competing interests.
References
1. Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib, Proc. Natl Acad Sci USA. 2004;101(36):13306–13311. doi:10.1073/pnas.0405220101
2. Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi:10.1126/science.1099314
3. Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi:10.1056/NEJMoa040938
4. Shi Y, Au JS-K, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER), J. Thorac Oncol. 2014;9(2):154–162. doi:10.1097/JTO.0000000000000033
5. Shigematsu H, Gazdar AF, et al. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int J Cancer. 2006;118(2):257–262. doi:10.1002/ijc.21496
6. Kuan FC, Kuo L-T, Chen M-C, et al. Overall survival benefits of first-line EGFR tyrosine kinase inhibitors in EGFR-mutated non-small-cell lung cancers: a systematic review and meta-analysis. Br J Canc. 2015;113(10):1519–1528. doi:10.1038/bjc.2015.356
7. Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362(25):2380–2388. doi:10.1056/NEJMoa0909530
8. Fong T, Morgensztern D, Govindan R, et al. EGFR inhibitors as first-line therapy in advanced non-small cell lung cancer. J Thorac Oncol. 2008;3(3):303–310. doi:10.1097/JTO.0b013e3181645477
9. Wu YL, Zhou C, Hu C-P, et al. Afatinib versus cisplatin plus gemcitabine for first line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised Phase 3 trial. Lancet Oncol. 2014;15(2):213–222. doi:10.1016/S1470-2045(13)70604-1
10. De Pas T, Toffalorio F, Manzotti M, et al. Activity of epidermal growth factor receptor-tyrosine kinase inhibitors in patients with non-small cell lung cancer harboring rare epidermal growth factor receptor mutations. J Thorac Oncol. 2011;6(11):1895–1901. doi:10.1097/JTO.0b013e318227e8c6
11. Yu HA, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19:2240–2247.
12. Li W, Qiu T, Guo L, et al. Primary and acquired EGFR T790M-mutant NSCLC patients identified by routine mutation testing show different characteristics but may both respond to osimertinib treatment. Cancer Lett. 2018;423:9–15. doi:10.1016/j.canlet.2018.03.005
13. Fujita Y, Suda K, Kimura H, et al. Highly Sensitive Detection of EGFR T790M Mutation Using Colony Hybridization Predicts Favorable Prognosis of Patients with Lung Cancer Harboring Activating EGFR Mutation. J Thorac Oncol. 2012;11(11):1640–1644. doi:10.1097/JTO.0b013e3182653d7f
14. Zhang B, Xu J, Zhang X, et al. Coexistence of sensitive and resistant epidermal growth factor receptor (EGFR) mutations in pretreatment non-small cell lung cancer (NSCLC) patients: first or third generation tyrosine kinase inhibitors (TKIs)? Lung Cancer. 2018;117:27–31. doi:10.1016/j.lungcan.2018.01.006
15. Ye X, Zhu -Z-Z, Zhong L, et al. High T790M Detection Rate in TKI-Naive NSCLC with EGFR Sensitive Mutation: truth or Artifact? J. Thorac Oncol. 2013;8(9):1118–1120. doi:10.1097/JTO.0b013e31829f691f
16. Ahn MJ, Lee JY, Lim SH, et al. Dynamic serial monitoring of EGFR mutations in plasma DNA samples in EGFR mutant NSCLC patients treated with EGFR TKI. J Clin Oncol. 2015;33(15_suppl):8078. doi:10.1200/jco.2015.33.15_suppl.8078
17. Watanabe M, Kawaguchi T, Isa S-I, et al. Ultra‑Sensitive detection of the pretreatment EGFR T790M mutation in non‑small cell lung cancer patients with an EGFR‑activating mutation using droplet digital PCR. Clin Cancer Res. 2015;21(15):
18. Inukai M, Toyooka S, Ito S, et al. Presence of Epidermal Growth Factor Receptor Gene T790M Mutation as a Minor Clone in Non–Small Cell Lung Cancer. Cancer Res. 2006;66(16):7854–7858. doi:10.1158/0008-5472.CAN-06-1951
19. Li Y, Xu Y, Wu X, et al. Comprehensive analysis of EGFR T790M detection by ddPCR and ARMS-PCR and the effect of mutant abundance on the efficacy of osimertinib in NSCLC patients. J Thorac Dis. 2019;11(7):3004–3014. doi:10.21037/jtd.2019.07.42
20. Wang S, Yan B, Zhang Y, et al. Different characteristics and survival in non-small cell lung cancer patients with primary and acquired EGFR T790M mutation. Int J Cancer. 2019;144(11):2880–2886. doi:10.1002/ijc.32015
21. Tatematsu T, Okuda K, Suzuki A, et al. The detectability of the pretreatment EGFR T790M mutations in lung adenocarcinoma using CAST-PCR and digital PCR. J Thorac Dis. 2017;9(8):2397–2403. doi:10.21037/jtd.2017.07.02
22. Panwen T, Wang Y, Wang W, et al. High-throughput sequencing reveals distinct genetic features and clinical implications of NSCLC with de novo and acquired EGFR T790M mutation. Lung Cancer. 2018;124:205–210. doi:10.1016/j.lungcan.2018.08.014
23. Hata AN, Niederst MJ, Archibald HL, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med. 2016;22(3):262–269. doi:10.1038/nm.4040
24. Hidaka N, Iwama E, Kubo N, et al. Most T790M mutations are present on the same, EGFR, allele as activating mutations in patients with non-small cell lung cancer. Lung Cancer. 2017;108:75–82. doi:10.1016/j.lungcan.2017.02.019
25. Iwama E, Takayama K, Harada T, et al. Highly sensitive and quantitative evaluation of the EGFR T790M mutation by nanofluidic digital PCR. Oncotarget. 2015;6(24):20466–20473. doi:10.18632/oncotarget.4058
26. Su KY, Chen H-Y, Li K-C, et al. Pretreatment Epidermal Growth Factor Receptor (EGFR) T790M Mutation Predicts Shorter EGFR Tyrosine Kinase Inhibitor Response Duration in Patients With Non–Small-Cell Lung Cancer. J Clin Oncol. 2012;30(4):433–440. doi:10.1200/JCO.2011.38.3224
27. Yang L, Sun L, Xiong Z-C, et al. Meta-analysis of the impact of de novo and acquired EGFR T790M mutations on the prognosis of, patients with non-small cell lung cancer receiving EGFR-TKIs</em>. OncoTargets Ther. 2017;10:2267–2279. doi:10.2147/OTT.S133082
28. Mulloy R, Ferrand A, Kim Y, et al. Epidermal growth factor receptor mutants from human lung cancers exhibit enhanced catalytic activity and increased sensitivity to gefitinib. Cancer Res. 2007;67(5):2325–2330. doi:10.1158/0008-5472.CAN-06-4293
29. Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR -Mutated Advanced Non–Small-Cell Lung Cancer. N Engl J Med. 2018;378(2):113–125. doi:10.1056/NEJMoa1713137
30. Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall Survival with Osimertinib in Untreated, EGFR -Mutated Advanced NSCLC, EGFR-Mutated Advanced NSCLC. N Engl J Med. 2020;382(1):41–50. doi:10.1056/NEJMoa1913662
31. Wang Z, Yang -J-J, Huang J, et al. Lung Adenocarcinoma Harboring EGFR T790M and In Trans C797S Responds to Combination Therapy of First- and Third- Generation EGFR TKIs and Shifts Allelic Configuration at Resistance. J Thorac Oncol. 2017;12(11):1723–1727. doi:10.1016/j.jtho.2017.06.017
32. Zheng D, et al. Association of plasma EGFR T790M ctDNA status with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep 6. 2016;20913:1–9.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

