Back to Journals » International Journal of Nanomedicine » Volume 16
Highly Efficient MicroRNA Delivery Using Functionalized Carbon Dots for Enhanced Conversion of Fibroblasts to Cardiomyocytes
Authors Yang L, Xue S, Du M, Lian F
Received 2 February 2021
Accepted for publication 1 May 2021
Published 1 June 2021 Volume 2021:16 Pages 3741—3754
DOI https://doi.org/10.2147/IJN.S304873
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Lei Yang
Lei Yang, Song Xue, Mingjun Du, Feng Lian
Department of Cardiovascular Surgery, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, 200127, People’s Republic of China
Correspondence: Feng Lian Email [email protected]
Introduction: The reprogramming of induced cardiomyocytes (iCMs) is of particular significance in regenerative medicine; however, it remains a great challenge to fabricate an efficient and safe gene delivery system to induce reprogramming of iCMs for therapeutic applications in heart injury. Here, we report branched polyethyleneimine (BP) coated nitrogen-enriched carbon dots (BP-NCDs) as highly efficient nanocarriers loaded with microRNAs-combo (BP-NCDs/MC) for cardiac reprogramming.
Methods: The BP-NCDs nanocarriers were prepared and characterized by several analytical techniques.
Results: The BP-NCDs nanocarriers showed good microRNAs-combo binding affinity, negligible cytotoxicity, and long-term microRNAs expression. Importantly, BP-NCDs/MC nanocomplexes led to the efficient direct reprogramming of fibroblasts into iCMs without genomic integration and resulting in effective recovery of cardiac function after myocardial infarction (MI).
Conclusion: This study offers a novel strategy to provide safe and effective microRNAs-delivery nanoplatforms based on carbon dots for promising cardiac regeneration and disease therapy.
Keywords: nitrogen-enriched carbon dots, branched polyethyleneimine, microRNAs, direct reprogramming, induced cardiomyocytes
Introduction
Heart failure, one of the most feared health conditions in humans, is attributed to hindered regenerative capability in cardiomyocytes and an impaired heart function.1,2 Inadequate regeneration of cardiomyocytes and excessive fibrosis are among the most familiar reasons for heart failure since this leads to substantial damage of tissue structure and function. In recent years, cell-based therapies, or the introduction of healthy cells into cardiac scar tissue, have modestly improved various measures of cardiac function, which has made major advances in cardiac regenerative medicine.3–5 However, concerns about the maturity and functional heterogeneity of stem cell-derived cardiomyocytes, the low survival, and retention of delivered cells, as well as their potential tumorigenicity still exist.6–8 To address these limitations, the reprogramming of induced-cardiomyocytes (iCMs) derived from abundant non-cardiomyocytes has been gaining tremendous attention as a novel therapy for cardiac diseases.9–11 Since the first reported three cardiac-lineage-related transcription factors, namely Gata4, Mef2c, and Tbx5 (GMT),12–14 different combinations of transcription factors and small molecules have also been identified to improve the effectiveness of cardiomyocyte reprogramming.15–18
An emerging therapeutic option for direct reprogramming of cardiac fibroblasts to cardiomyocytes involves microRNAs (miRNAs). miRNAs play an essential role in regulating biological processes, including controlling gene transcription through the activation or degradation of targeted mRNA. Some preeminent works have demonstrated miRNAs-combo as highly efficient reprogramming tools. For instance, Jayawardena et al provided proof-of-concept that miRNAs have the capability of directly converting fibroblasts to a cardiomyocyte-like phenotype in vitro and cardiac reprogramming in vivo.22 The repressive actions of miRNAs on gene expression can be powerful as a single miRNA may target multiple pathways simultaneously.19–21 Chemically synthesized miRNAs mimics are easily administered to cells via lipid-based transfection and exhibit low toxicity in experiments.22,23 Moreover, small size of a single miRNA allows the packing of multiple transcripts in the same delivery vector to increase reprogramming efficiency and functional homogeneity of reprogrammed cells.24 However, administering naked miRNAs is difficult based on its surface charge (negatively charged) and potential rapid degradation. Therefore, highly efficient and biocompatible vehicles are necessary to protect and deliver miRNAs into targeted cells.
Viruses (eg, retroviruses, and lentiviruses) and liposomes have been widely studied as gene vectors to convert fibroblasts to cardiomyocytes directly.4,12,15,22,25 However, disadvantages such as genotoxic transgene integration and the ability to trigger immune responses, which may lead to insertable mutations and tumor formation, have prevented its therapeutic potential.26 Thus, novel non-viral reprogramming systems are required to allow the clinical application of cardiac reprogramming.24,27–31 Non-viral vector systems including cationic lipids, cationic polymers, dendrimers, and peptides have shown good biocompatibility and the potential for systemic gene delivery.32–35 Artificial cationic polymers PEI with an MW of 25 kDa is acknowledged as the “gold standard” of polymeric gene vectors for its excellent gene transfection efficiency and universality. Although the higher charge density of cationic polymers PEI provides strong nucleic acid binding ability, these polyplexes are confronted with the problem of cytotoxicity.36 These barriers have motivated researchers to find new strategies to promote the performance of non-viral gene vectors, and simultaneously allow the materials to be multifunctional to meet disease therapy requirements.
Carbon dots (CDs) have drawn intensive interest in the nanotechnology field due to their high water solubility, favorable photoluminescence property, good chemical stability, multifunctional surface functions, and low cytotoxicity.37–42 To explore their delivering capability for potential therapy, much work has been focused on the surface modification of CDs with biocompatible ligands (ie, biomacromolecules, peptides, antibodies, etc.).43–45 Although CDs have been successfully studied in biosensor, cell imaging, and recognition fields, their applications as non-viral vectors with directly transformed reprogramming factors have rarely been reported.
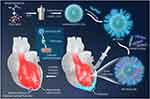
Herein, we designed branched polyethyleneimine modified N-enriched CDs (BP-NCDs) with low toxicity for use as a non-viral vector. Then, loaded BP-NCDs with miRNAs-combo via electrostatic interactions to form targeted nanocomplexes (BP-NCDs/MC). We found that reprogramming fibroblasts into iCMs using BP-NCDs/MC nanocomplexes resulted in a significant upregulation of cardiomyocyte genes and a gradual inhibition of fibroblast genes. Furthermore, local injection of BP-NCDs/MC nanocomplexes into mouse hearts significantly improved cardiac function after myocardial infarction (Figure 1). This study not only shows the versatility of BP-NCDs/MC nanocomplexes as delivery vectors to directly reprogram fibroblasts into iCMs without genomic integration but also offers inspiration for the exploration of applications of CD-based gene/drug delivery systems in cardiac injury treatment.
Materials and Methods
Methods
Preparation of BP-NCDs/MC nanocomplexes and agarose gel electrophoresis. BP-NCDs solution was prepared with ultra-pure water. After lyophilized, the solution was weighed and standardized BP-NCDs (1 mg/mL). The solution was then sterilized with a 0.22μm filter screen before use. Four miRNA mimics: miRNA-1, miRNA-133, miRNA-208, and miRNA-499 were used to prepare BP-NCDs/MC nanocomplexes. Predetermined concentrations of sterilized BP-NCDs were sonicated for 15 min, then added into the defined miRNAs-combo solution at different weight ratios used vortex for assembly. The obtained BP-NCDs/MC nanocomplexes were static at room temperature for 15 min and then stored at 4°C for subsequent use. Next, BP-NCDs/MC nanocomplexes (miRNA-1: miRNA-133: miRNA-208: miRNA-499 = 1:1:1:1:1) was selected for agarose gel electrophoresis. The nanocomplexes (10 μL) solution was added to 2 μL of loading buffer (0.1% sodium dodecyl sulfate, 5% glycerol, and 0.005% bromophenol blue) and applied to a 2% agarose gel in a TAE acid buffer solution (pH 8.0) containing 0.1 mg/mL ethidium bromide (EtBr). The complex was subjected to electrophoresis in TAE solution at a voltage of 120 V for 30 min. The gel is imaged with a UV diopter.
Cell Culture
Cardiac fibroblasts (CFs) were extracted from the C57BL/6 mice on 3 days of birth. Hearts were removed from the chest of mice and rinsed with fresh PBS. Tissues were decomposed with ophthalmic scissors, and then incubated in 0.08% trypsin (4.5 mL) and 0.08% collagenase II (10 mg collagenase II dissolved in the 10 mL pure medium) for 45 minutes. After 3–4 times digestion, the cells were collected by differential centrifugation and placed in a T75 culture flask containing high glucose DMEM (10% FBS and 1% penicillin/streptomycin). The cells passed to the third generation were transfected into BP-NCDs, BP-NCDs/MC, and PEI/MC complexes in a 6-well culture plate. The transfected cells were cultured in an induction medium (10% FBS and 1% penicillin/streptomycin). Replace the medium with the transforming growth factor-β inhibitor SB431542 and the WNT inhibitor XAV939 after 24h and replace the medium every 48h.16
Cell Viability Assays
Cardiac fibroblasts (1.0×104) were cultured in 96-well plates with high glucose DMEM (10% FBS and 1% penicillin/streptomycin) for 12h. Fresh serum-free medium (Opti-MEM) was added and incubated with BP-NCDs, PEI/MC, and BP-NCDs/MC for 6h. A quantity of 10μL CCK-8 solution was added to each well and incubated at room temperature for another 4h. The optical density (OD) of the mixture was measured at 450 nm with a microplate reader. Cardiac fibroblasts (20.0×104) were placed in 6-well culture plates and transfected with BP-NCDs, PEI/MC, and BP-NCDs/MC nanocomplexes for 6h. To remove the supernatant, wash the cells, make sure to remove the active esterase in the culture medium, add a sufficient amount of the Live & Dead Viability Assay Kit working liquid, incubate at room temperature for 30–45 minutes, absorb the dyeing working fluid to stop incubation, wash the PBS, observe the labeled cells under the fluorescence microscope.
Cellular Uptake Efficiencies
The transfection efficiency was measured by fluorescence confocal microscopy and flow cytometry. Cardiac fibroblasts were transfected into Opti-MEM medium using BP-NCDs, PEI/MC, and BP-NCDs/MC. The blank group was used as the control group. Cardiac fibroblasts were seeded in a polylysine-coated confocal dish and cultured at 37°C for 12h. When the cells adhered to the surface of the culture, the cardiac fibroblasts were transfected with Cy5-labeled miRNA compound in Opti-MEM medium. After incubation at 37°C for 6h, the cells were washed with PBS, then observed using a fluorescence confocal microscopy. Flow cytometry was used to monitor the quantitative cellular uptake of nanoparticles in cardiac fibroblasts. Cardiac fibroblasts were transfected with Cy5-labeled miRNA nanocomplexes for 6h, then washed with PBS, followed by trypsinization, centrifugation, resuspended in 200 μL of PBS containing 2% fetal bovine serum, added with 200 μL of 4% paraformaldehyde, and incubated at 4°C for 2h. The fluorescence intensity distribution of 10,000 cell samples was measured and analyzed using FlowJo software.
Bright Field Imaging
After transfection, the cardiac fibroblasts were cultured for 3 weeks and observed under a bright field microscope. After washing with PBS twice, the cells were observed under a Leica fluorescence microscope, and iCM cells were counted.
Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction
Total RNA was extracted from cardiac fibroblasts via BP-NCDs, PEI/MC, or BP-NCDs/MC complexes using standard methods and PCR using primers. The RNA was collected and purified using the EZ-Press RNA Purification Kit (EZBioscience, Fairview Ave, USA) according to the manufacturer’s instructions. The total RNA concentration was quantified using a NanoDrop spectrophotometer. RNA was reverse-transcribed into cDNA using PrimeScrip RT-PCR Kit (TaKaRa, Shiga, Japan). The quantitative real-time PCR reaction was performed using a LightCycler from Roche Molecular Biochemicals, using SYBR premixed Ex Taq (TaKaRa, Shiga, Japan), according to the protocol provided by the manufacturer. The PCR reaction process was as follows: denaturation at 95°C for 10 min (initial denaturation) followed by 30 cycles, denaturation at 95°C for 30 s, then annealing at 57°C for 30 s, and elongation at 72°C for 30 s. GAPDH is used as an internal standard. A no template blank and a reverse transcriptional negative blank served as a negative control. 2∧(-∆∆Ct) method was used to process the data. Gene expression of each factor was normalized by GAPDH. All the measurements were made in triplicates. The sequence of target primers is shown in (Table S1).
Western Blotting
After 1 and 2 weeks of transfection, the cells were washed by PBS and digested by trypsin. The cells obtained were lysed through centrifugation in an ice bath with RIPA lysis buffer containing protease inhibitor for 15 min, followed by ultrasonic treatment for 5 min, centrifuged at 12,000 g for 15 min, and the supernatant collected. The protein concentration was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL, USA), and then 5×SDS-PAGE non-reducing protein loading buffer was added and boiled at 95° for 10 min. Equivalent protein samples were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membrane. The membranes were blocked using 5% BSA and incubated with primary antibodies against Gata4 (1:2000; Cambridge, UK, Abcam), Mef2c (1:1000; Cambridge, UK, Abcam), Tbx5 (1:1000; Cambridge, UK, Abcam), Hand2 (1:1000; Cambridge, UK, Abcam) and GAPDH (1:500; Cambridge, UK, Abcam). Then, the membrane was incubated with Goat anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 second incubation for 1h at room temperature, using the LI-COR company’s Odyssey two-color infrared fluorescence imaging system scans.
Animals
C57BL/6 mice were provided by and housed in an animal facility at Renji Hospital under specific pathogen-free (SPF) condition. All animal procedures were performed in accordance with NIH guidelines (Guide for the care and use of laboratory animals) and approved by Institutional Animal Care and Use Committee of Shanghai Jiao Tong University School of Medicine (License: SYXK (Shanghai) 2016–0009, Ethic code: A2020124).
MI Induction and miRNA-Combo Delivery in Infarcted Hearts
In this study, MI was induced by permanent ligation of the left anterior descending artery (LAD). To deliver the miRNA complex to the infarcted heart tissue, a pool of miRNA mimics was prepared, and 20 μL of PEI/MC, BP-NCDs/MC nanocomplexes were injected into the epicardium along the boundary between the infarcted area and the boundary area. Four weeks after MI and injection of miRNA complex, the heart was retrogradely perfused with PBS containing 4% PFA through the right carotid artery. Paraffin-embedded tissue sections (Torrance, CA, Sakura Finetek) were then performed. Infarct size (percentage of fibrosis length and infarct thickness) was shown by Masson staining. The heart sections were observed under a light microscope (Tokyo, Japan, Olympus). The area of fibrotic tissue was then measured using ImageJ software version 1.8.0 (National Institutes of Health) and expressed as the percentage of the fibrosis area and infarct thickness.
Immunofluorescence Staining
After transfection of PEI/MC and BP NCDs/MC nanocomplexes, the digested cell suspension was transferred to the confocal culture dish. After PBS washing, 4% polyformaldehyde fixed 1h, after PBS washing, with 0.2% of Triton X-100 rupture in 1h at room temperature, with PBS washing three times, add a goat serum albumin (GSA, Shanghai, China, Beyotime), down closed 2h at room temperature, out of confining liquid washing after adding suitable amount of COL1A1 (1:200; Cambridge, UK, Abcam), cTnI (1:500; Cambridge, UK, Abcam). The antibodies in each petri dish were then placed in a wet box and incubated overnight at 4°C. And then incubated with secondary antibodies (Goat Anti-Mouse IgG H&L (Alexa Fluor 488); Goat Anti-Mouse IgG H&L (Alexa Fluor 647), Cambridge, UK, Abcam) and 4ʹ,6-diamidino-2- phenylindole (DAPI) for 1h at room temperature. After washed with TBST (Shanghai, China, Beyotime), the cells were visualized under a Nikon fluorescence microscope.
For immunohistochemistry, heart tissues were stained with CD31 (1:200; Cambridge, UK, Abcam) for 24h at 4°C and then incubated with secondary antibodies (Goat Anti-Mouse IgG H&L (Alexa Fluor 647), Cambridge, UK, Abcam) and DAPI for 1h at room temperature. Sections were observed under a universal Nikon fluorescence microscope.
Statistical Analysis
All data were expressed as mean ± SEM of three independent experiments. The n value represents the number of independent experiments conducted, the number of individual experiments, or the number of mice. P < 0.05 was considered significant (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). One-way analysis of variance (ANOVA) was used for the analysis of significant differences between groups. After confirming the normal distribution, Tukey multiple-comparison test and Student’s t-test were used for the double-component comparison. Prism GraphPad Software was used for statistical analysis.
Results and Discussion
Synthesis and Characterization of BP-NCDs/MC Nanocomplexes
The synthesis of BP-NCDs/MC nanocomplexes involved three steps: (i) synthesis of N-doped CDs (N-CDs), (ii) grafting branched PEI on the surface of carboxyl-capped N-CDs (BP-NCDs), and (iii) construction of BP-NCDs/MC nanocomplex through the electrostatic interactions between positive-charged BP-NCDs and negative-charged miRNAs-combo (miRNA-1, miRNA-133, miRNA-208, and miRNA-499) (Figure 1). Transmission electron microscope (TEM) and Atomic force microscopy (AFM) images give the morphological information of N-CDs, and BP-NCDs. N-CDs exhibited a mean diameter of 30.5 nm by TEM analysis (Figure 2A). The size of BP-NCDs was mainly about 41.0 nm, showing a 10.5 nm increase compared to the bare N-CDs (Figure 2B). Meanwhile, the functionalization of BP-NCDs led to a height increase from 0.42 nm to 0.90 nm by the AFM analysis (Figure 2C and D). The average hydrodynamic size of the BP-NCDs measured by DLS was 42.1 nm, which was slightly larger than that of the bare N-CDs (30.4 nm) due to the presence of the polymer PEI envelope (Figure 2E). 47 The surface charge of BP-NCDs reflects the capability of cationic-related carriers for nucleic acid delivery.43 Zeta potential analysis demonstrated that the surface charge of N-CDs, branch PEI, and BP-NCDs was 7 mV, 10 mV, and 18 mV, respectively (Figure 2F). These results together suggest that branched PEI was successfully grafted on the surface of the N-CDs.44–47
The X-ray-photoelectron spectroscopy (XPS) measurements were carried out to elucidate the difference in the elemental composition of pure N-CDs and BP-NCDs. The full scan of XPS spectra showed three prominent peaks at 284 eV, 401 eV, and 532 eV attributed to C1s, N1s, and O1s, respectively. The relative elemental analysis showed an increase of carbon and nitrogen levels in BP-NCDs, with an associated decrease in oxygen content (67.34 at % C1s, 17.10 at % N1s and 15.56 at % O1s) when compared to the N-CDs (53.02 at % C1s, 8.53 at % N1s and 38.45 at. % O1s) (Figure 3A). In the high-resolution C1s spectrum of N-CDs, three peaks attribute to C-C or C=C (sp2, 284.4 eV), C-N, or C-O (sp3, 285.2 eV), C=O (sp3, 287.5 eV) bonds were observed. However, the high-resolution C1s spectrum of BP-NCDs showed four peaks, presumably due to the formation of amide groups between the carboxylic group of N-CDs and the amine group of branched PEI (Figure 3B). The two main peaks of the N1s spectrum for BP-NCDs were associated with the pyridinic N (399.5 eV) and the quaternary N (401.5 eV). Simultaneously, the two main peaks of the N1s spectrum for N-CDs was linked to the pyrrolic N (399.9 eV) and quaternary N (401.8 eV) (Figure 3C). This change is likely due to the formation of the amide bonds, which is in good agreement with the C1s XPS spectrum.46 The functional groups and chemical compositions of N-CDs and BP-NCDs were further identified by Fourier transform infrared (FT-IR) spectra (Figure 3D). N-CDs revealed a broad peak at 3408 cm-1, which confirms the existence of hydroxyl(-OH) groups from the carboxylic groups and water molecules or N–H stretching from the amine structure. The sharp peak at 1698 cm-1 is typical for carbonyl (C=O) groups. Specifically, the band at 1400 cm-1 showed the stretching and bending vibrations of the C-C bond, and the peak at 1122 cm-1 was ascribed to C-O stretching, which is often found in the oxidized carbons. While a broad peak loaded at 3338 cm-1 was attributed to the -OH group and carboxyl group of BP-NCDs. The apparent absorption amide I (1636 cm−1) and amide bands II (1573 cm−1) peaks corresponded to the amide of -CONH- groups in the PEI functionalized N-CDs. The crystallinity of the bare N-CDs was examined by X-ray diffraction (XRD). The XRD pattern exhibited a broad peak between 20° and 35° 2θ, which is attributed to the graphitic structure of carbon47 (Figure S1).
In the UV-vis experiment, the maximum absorption peak of N-CDs was located at 345 nm. After the N-CDs were functionalized with branched PEI, the band of BP-NCDs blue-shifted to 270 nm. The inset in Figure 3E are photographs of bare N-CDs and BP-NCDs solutions under a visible light. An excitation-dependent fluorescence spectrum of N-CDs was observed firstly, and 350 nm is the optimal excitation wavelength (Figure S2). The bare N-CDs showed a strong fluorescence spectrum peak at 440 nm when excited at 350 nm. The maximum emission peak of BP-NCDs red-shifted to 445 nm, and the inset in Figure 3F are photographs of bare N-CDs and BP-NCDs solutions under UV light (365 nm). These results suggested that the surface passivation by PEI imparted a strong influence on the optical properties of N-CDs due to a change in the chemical functional groups.47 Furthermore, the time-resolved fluorescence spectrum measured with an excitation at 350 nm showed that the fluorescence lifetime of N-CDs is 3.35 ns (Figure S3).
Agarose Gel-Retardation Study
Agarose gel delay studies were conducted under different BP-NCDs/miRNA weight ratios to test the potential of BP-NCDs as efficient gene delivery vectors. The mass ratio of BP-NCDs/miRNAs was set as 1:1, 2:1, 3:1, 4:1, and 5:1. The corresponding results showed that when the mass ratio of BP-NCDs/miRNAs was increased to 4:1, miRNAs remained completely in the original pore without simulating the migration of miRNAs in the gel (Figure 4A). This finding indicated that BP-NCDs has an excellent complexing ability with miRNAs-combo due to its good positive surface charge.
Cell Viability Assay
The cytotoxicity of BP-NCDs and BP-NCDs/MC nanocomplexes with the same miRNAs-combo weight was examined by CCK-8 method. The PEI/MC group was used as a positive control. The measured absorbance is normalized to the absorbance of the untreated cells. The results showed that the viability of cardiac fibroblasts culture in BP-NCDs and BP-NCDs/MC nanocomplexes was higher than that in PEI/MC group (BP-NCDs/miRNAs mass ratio of 4:1) (Figure 4B). Twenty-four hours after transfection of the same mass of miRNAs in the growth medium, the Live/Dead assay (Calcein AM) indicated excellent cell viability associated with the BP-NCDs/MC group compared to the BP-NCDs and PEI/MC groups (Figure S4). This result suggested that small-sized BP-NCDs have good biocompatibility in gene delivery.
Transfection and Delivery of miRNAs-Combo by BP-NCDs
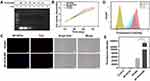
We evaluated the ability of BP-NCDs as gene vectors using functionalized fluorescent carbon dots constructs. Blue fluorescence was observed from the intracellular BP-NCDs (Figure 4C, blue field). After miRNAs-combo hybridized with intracellular target mRNA, the fluorescent dye was isolated from the quencher and dissociated from the duplex structure on the surface of BP-NCDs to produce red fluorescence (Figure 4C, red field). The quantification of the extent of nanoparticle internalization by blue and red fields coverage and fluorescence-activated cell sorting (FACS) further confirmed that BP-NCDs cell uptake is higher than PEI (Figure 4D and E). These results indicated that miRNAs-combo were effectively transferred into cells, which led to the up-regulation of intracellular miRNAs levels. The multifunctional BP-NCDs designed in this work are efficient in cell gene regulation and fluorescence imaging.
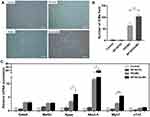
BP-NCDs Nanocomplexes Directly Induced Cardiac Fibroblasts to be Reprogrammed Into Functional iCMs in vitro
We examined the presence of cell colony morphology in BP-NCDs, PEI/MC, and BP-NCDs/MC groups using iCMs. BP-NCDs/MC group had a significant amount of iCMs within 3 weeks, and we observed that the transduction of BP-NCDs/MC to iCMs produced higher contractile activity as well as more iCMs than the BP-NCDs group (Figure 5A). The expression of iCMs gene in cells transfected with BP-NCDs/MC nanocomplex was measured to demonstrate the effect of miRNA transfection on the direct reprogramming of myocardial fibroblasts at the mRNA level (Figure 5C). In carrying out this experiment, specific genes during myocardial development as well as maturation, ie, GATA Binding Protein 4 (GATA4), Myocyte enhancer factor 2C (MEF2C), Natriuretic Peptide A (NPPA), Myosin Heavy Chain 7 (MYH7), NK2 Homeobox 5 (NKX2-5), Troponin T2, Cardiac Type (TNNT2) were selected. On day 7, the iCMs gene expression in the BP-NCDs group was not significantly different from that of the control group. The cells transfected with PEI/MC and the cells transfected with BP-NCDs/MC nanocomplex were elevated considerably in myocardial-specific gene expression relative to the control cells. Regarding the expression of the genes (ie, GATA4, MEF2C, NPPA, NKX2-5) in the early stage of cardiac development, the cells transfected with PEI/MC nanocomplex were almost no different from those transfected with BP-NCDs/MC. In contrast, the cells transfected with BP-NCDs/MC nanocomplex were higher than those transfected with PEI/MC in the expression of genes (MYH7, TNNT2) in the late maturation of the heart.
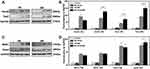
Transcription factors in early cardiac development are crucial to the successful reprogramming of fibroblasts to iCMs,8 and the high expression of cardiac transcription factors Gata4, Hand2, Mef2c, and Tbx5 proteins has been shown to transform fibroblasts into cardiac progenitors effectively.46 We used Western blotting to detect proteins that play an important role in the reprogramming process, including GATA Binding Protein 4 (Gata4), Myocyte Enhancer Factor 2C (Mef2c), T-Box Transcription Factor 5 (Tbx5), and Heart and Neural Crest Derivatives Expressed 2 (Hand2). As shown after 7 and 14 days of incubation in BP-NCDs, PEI/MC, BP-NCDs/MC, and the control groups, myocardial development-related proteins were not expressed. The expression levels of myocardial development-related proteins were significantly up-regulated in the PEI/MC and BP-NCDs/MC groups. Moreover, the expression level of BP-NCDs/MC group was higher than that of the PEI/MC group (Figure 6A and B). In summary, our results showed that both the PEI/MC and BP-NCDs/MC groups significantly increased the expression of Gata4, Mef2c, Tbx5, Hand2, which was consistent with our real-time PCR results.
Cardiac fibroblasts have the potential to differentiate, but in vitro, the function of iCMs derived from cardiac fibroblasts is not mature. Since transdifferentiating is thought to be a long and random process, it would be interesting to produce a fully reprogrammed iCM. In previous studies, cardiac fibroblasts had been reprogrammed to resemble cardiac muscle cells after being maintained in the induction medium for 4 to 11 weeks.11–20 In this study, cardiac fibroblasts were transfected with PEI/MC and BP-NCDs/MC and cultured in the induction medium for 4 weeks. And through immunofluorescence staining, we observed that differentiated cells expressed iCMs marker proteins such as cTnI (Figure 7). In other reports, fibroblast genes were gradually inhibited during the reprogramming of iCMs.47–50 miRNA-133 direct target Snai1 3 ʹ UTR and restrain fiber mother cell surface markers such as COL1A1, COL1A2, POSTN, promote fibroblasts transition to the heart muscle cells.21 With the reprogramming, the expression of COL1A1 decreases over time. Therefore, these results indicated that BP-NCDs/MC nanocomplexes significantly enhanced the expression of myocardial development and related proteins. The binding gene expression results showed that the functionalized BP-NCDs/MC nanocomplexes delivered a significant process of promoting cardiac fibroblasts to iCMs. These results suggest that cardiac reprogramming initiates iCMs reprogramming by activating endogenous GHMT genes while inhibiting fibrosis pathways. Therefore, fibrosis may be a major obstacle to cardiac reprogramming, and future research should focus on effective suppression of fibrosis.19–21
BP-NCDs/MC Nanocomplexes Improves the Treatment of Myocardial Infarction in vivo
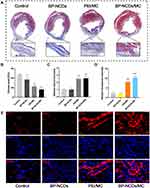
Related studies have shown that after viral gene transfer, fibrosis is reduced in the body to improve heart function.14 Therefore, we proposed that BP-NCDs/MC nanocomplexes could effectively promote myocardial infarction (MI) damage repair. To verify this, we induced cardiac damage by coronary artery ligation and injection of BP-NCDs/MC nanocomplexes into the myocardium at the infarct junction of a mouse heart. After 4 weeks of transduction, we evaluated the efficacy of BP-NCDs/MC nanocomplex in the mice compared to the control group (ie, untreated group). Surprisingly, the infarct area and scar area of MI mice treated with BP-NCDs/MC nanocomposite were significantly reduced compared to the control group (Figure 8A). Besides, in the BP-NCDs/MC nanocomplexes treatment group, the fibrotic area and infarct thickness significantly decreased (Figure 8B and C). Immunohistochemical examination was performed 4 weeks later, with histological sections taken at the infarct site, and CD31 was used as a marker of cardiac regeneration to assess fibrosis. In the PEI/MC and BP-NCDs/MC treatment group, CD31 were significantly increased compared to the control and BP-NCDs group (Figure 8D and E).
Conclusion
In summary, the surface of N-CDs was successfully modified using branched polyethyleneimine (BP, 25 kDa), resulting in BP-NCDs, which can be used as non-viral vectors for miRNAs-combo (miRNA-1, 133, 208, and 499) delivery. The BP-NCDs nanovector showed high miRNAs-combo binding affinity, stable expression in cells, excellent cellular uptake, and low levels of cytotoxicity. Additionally, BP-NCDs/MC nanocomplexes significantly enhanced the direct reprogramming of fibroblasts into iCMs without genomic integration, and the injection of BP-NCDs/MC nanocomplexes into mouse infarcted hearts significantly improved the quality, and quantity of cardiac functions after myocardial infarction (MI). Thus, our results suggest that BP-NCDs/MC nanocomplexes-mediated cardiac reprogramming might improve the safety and efficiency of therapeutic in heart injury. Although direct transformation induced by the non-viral vector remains a challenge and has a long way to go before clinical applications, this work may provide a strong support for further developing transdifferentiating techniques for clinical applications.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81873461).
Disclosure
The authors report no conflicts of interest.
References
1. Zhao D, Liu J, Wang M, et al. Epidemiology of cardiovascular disease in China: current features and implications. Nat Rev Cardiol. 2019;16(4):203–212. doi:10.1038/s41569-018-0119-4
2. Hausenloy DJ, Yellon DM. Myocardial ischemia-reperfusion injury: a neglected therapeutic target. J Clin Invest. 2013;123:92–100. doi:10.1136/heartjnl-2015-307855
3. Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi:10.1016/j.cell.2007.11.019
4. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi:10.1016/j.cell.2006.07.024
5. Nelson TJ, Martinez-Fernandez A, Yamada S, et al. Repair of Acute myocardial infarction by human stemness factors induced pluripotent stem cells. Circulation. 2009;120:408–416. doi:10.1161/CIRCULATIONAHA.109.865154
6. Mummery C. Induced pluripotent stem cells — a cautionary note. N Engl J Med. 2011;364:2160–2162. doi:10.1056/NEJMcibr1103052
7. Nishimori M, Yakushiji H, Mori M, et al. Tumorigenesis in cells derived from induced pluripotent stem cells. Cell. 2014;27:29–35. doi:10.1007/s13577-013-0078-3
8. Yamashita T, Kawai H, Tian F, et al. Tumorigenic development of induced pluripotent stem cells in ischemic mouse brain. Cell Transplant. 2011;20:883–892. doi:10.3727/096368910X539092
9. Srivastava D, Ivey KN. Potential of stem-cell-based therapies for heart disease. Nature. 2006;441:1097–1099. doi:10.1038/nature04961
10. Passier R, van Laake LW, Mummery CL. Stem-cell-based therapy and lessons from the heart. Nature. 2008;453:322–329. doi:10.1038/nature07040
11. Ieda M, Fu JD, Delgado-Olguin P, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi:10.1016/j.cell.2010.07.002
12. Qian L, Berry EC, Fu JD, et al. Reprogramming of mouse fibroblasts into cardiomyocyte-like cells in vitro. Nat Protoc. 2013;8:1204–1215. doi:10.1038/nprot.2013.067
13. Qian L, Huang Y, Spencer CI, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi:10.1038/nature11044
14. Song K, Nam YJ, Luo X, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi:10.1038/nature11139
15. Islas JF, Liu Y, Weng KC, et al. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci. 2012;109:13016–13021. doi:10.1073/pnas.1120299109
16. Mohamed TM, Stone NR, Berry EC, et al. chemical enhancement of in vitro and in vivo direct cardiac reprogramming. Circulation. 2017;135:978–995. doi:10.1161/CIRCULATIONAHA.116.024692
17. Zhao Y, Londono P, Cao Y, et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat Commun. 2015;6:8243. doi:10.1038/ncomms9243
18. Zhou H, Dickson ME, Kim MS, et al. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc Natl Acad Sci. 2015;112:11864–11869. doi:10.1073/pnas.1516237112
19. Dal-Pra S, Hodgkinson CP, Mirotsou M, et al. Demethylation of H3K27 is essential for the induction of direct cardiac reprogramming by miR combo. Circ Res. 2017;120:1403–1413. doi:10.1161/CIRCRESAHA.116.308741
20. Liu Z, Chen O, Zheng M, et al. Re-patterning of H3K27me3, H3K4me3 and DNA methylation during fibroblast conversion into induced cardiomyocytes. Stem Cell Res. 2016;16:507–518. doi:10.1016/j.scr.2016.02.037
21. Muraoka N, Yamakawa H, Miyamoto K, et al. MiR‐133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014;33:1565–1581. doi:10.15252/embj.201387605
22. Jayawardena TM, Egemnazarov B, Finch EA, et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110:1465–1473. doi:10.1161/CIRCRESAHA.112.269035
23. Yoo AS, Sun AX, Li L, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi:10.1038/nature10323
24. Miyoshi N, Ishii H, Nagano H, et al. Reprogramming of mouse and human cells to pluripotency using mature MicroRNAs. Cell Stem Cell. 2011;8:633–638. doi:10.1016/j.stem.2011.05.001
25. Maherali N, Ahfeldt T, Rigamonti A, et al. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–345. doi:10.1016/j.stem.2008.08.003
26. Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi:10.1038/nature05934
27. Carey BW, Markoulaki S, Hanna JV, et al. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc Natl Acad Sci. 2009;106:157–162. doi:10.1073/pnas.0811426106
28. Kim D, Kim CH, Moon JI, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi:10.1016/j.stem.2009.05.005
29. Judson RL, Babiarz JE, Venere M, et al. Embryonic stem cell–specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–461. doi:10.1038/nbt.1535
30. Cao X, Deng W, Qu R, et al. Non‐Viral Co‐Delivery of the Four Yamanaka Factors for Generation of Human Induced Pluripotent Stem Cells via Calcium Phosphate Nanocomposite Particles. Adv Funct Mater. 2013;23:5403–5411. doi:10.1002/adfm.201203646
31. Mormone E, Sousa SD, Alexeeva V, et al. “Footprint-free” human induced pluripotent stem cell-derived astrocytes for in vivo cell-based therapy. Stem Cells Dev. 2014;23:2626–2636. doi:10.1089/scd.2014.0151
32. Midoux P, Breuzard G, Gomez JP, et al. Polymer-based gene delivery: a current review on the uptake and intracellular trafficking of polyplexes. Curr Gene Ther. 2008;8:335–352. doi:10.2174/156652308786071014
33. Deng WW, Cao X, Wang M, et al. Efficient gene delivery to mesenchymal stem cells by an ethylenediamine‐modified polysaccharide from mulberry leaves. Small. 2012;8:441–451. doi:10.1002/smll.201101554
34. Das P, Bhattacharyy SK, Banerjic P, et al. Acoustic cavitation assisted synthesis and characterization of photoluminescent carbon quantum dots for biological applications and their future prospective. Nano-Struct Nano Objects. 2021;25:100641. doi:10.1016/j.nanoso.2020.100641
35. Bhattacharyya S, Dule M, Paul R, et al. Carbon dot cross-linked gelatin nanocomposite hydrogel for pH-Sensing and pH-responsive drug delivery. ACS Biomater Sci Eng. 2020;6(10):5662–5674. doi:10.1021/acsbiomaterials.0c00982
36. Essex S, Navarro G, Sabhachandani P, et al. Phospholipid-modified PEI-based nanocarriers for in vivo siRNA therapeutics against multidrug-resistant tumors. Gene Ther. 2015;22:257–266. doi:10.1038/gt.2014.97
37. Xia C, Zhu S, Feng T, et al. Evolution and synthesis of carbon dots: from carbon dots to carbonized polymer dots. Adv Sci. 2019;23:1901316. doi:10.1002/advs.201901316
38. Chung S, Revia RA, Zhang M. Graphene quantum dots and their applications in bioimaging, biosensing, and therapy. Adv Mater. 2019;12:1904362. doi:10.1002/adma.201904362
39. Yang S, Wang X, He P, et al. Carbon quantum dots and their applications. Chem Soc Rev. 2015;44:362–381. doi:10.1002/smll.202004867
40. Liu C, Zhang P, Zhan XY, et al. Nano-carrier for gene delivery and bioimaging based on carbon dots with PEI-passivation enhanced fluorescence. Biomaterials. 2012;33:3604. doi:10.1016/j.biomaterials.2012.01.052
41. Pierrata P, Wang R, Kereselidzea D, et al. Biomaterials. 2015; 51: 290. doi:10.1016/j.biomaterials.2015.02.017
42. Kim S, Choi Y, Park G, et al. Highly efficient gene silencing and bioimaging based on fluorescent carbon dots in vitro and in vivo. Nano Res. 2017;10(2):503–519. doi:10.1007/s12274-016-1309-1
43. Verma A, Stellacci F. Effect of surface properties on nanoparticle–cell interactions. Small. 2010;6:12–21. doi:10.1002/smll.200901158
44. Nekoueian K, Amiri M, Sillanpää M, et al. Carbon-based quantum particles: an electroanalytical and biomedical perspective. Chem Soc Rev. 2019;48:4281–4316. doi:10.1039/c8cs00445e
45. Chen J, Wang Q, Zhou J, et al. Porphyra polysaccharide-derived carbon dots for non-viral co-delivery of different gene combinations and neuronal differentiation of ectodermal mesenchymal stem cells. Nanoscale. 2017;9:10820–10831. doi:10.1039/c7nr03327c
46. Chang Y, Lee E, Kim J, et al. Efficient in vivo direct conversion of fibroblasts into cardiomyocytes using a nanoparticle-based gene carrier. Biomaterials. 2019;192:500–509. doi:10.1016/j.biomaterials.2018.11.034
47. Shu M, Gao F, Yu C, et al. Dual-targeted therapy in HER2-positive breast cancer cells with the combination of carbon dots/HER3 siRNA and trastuzumab. Nanotechnology. 2020;31:335102. doi:10.1088/1361-6528/ab8a8a
48. Li XH, Li Q, Jiang L, et al. Generation of functional human cardiac progenitor cells by high‐efficiency protein transduction. Stem Cells Transl Med. 2015;4:1415–1424. doi:10.5966/sctm.2015-0136
49. Liu Z, Wang L, Welch JD, et al. Single-cell transcriptomics reconstructs fate conversion from fibroblast to cardiomyocyte. Nature. 2017;551:100–104. doi:10.1038/nature24454
50. Lalit PA, Salick MR, Nelson DO, et al. Lineage reprogramming of fibroblasts into proliferative induced cardiac progenitor cells by defined factors. Cell Stem Cell. 2016;18:354–367. doi:10.1016/j.stem.2015.12.001
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.