Back to Journals » OncoTargets and Therapy » Volume 11
High SRPX2 protein expression predicts unfavorable clinical outcome in patients with prostate cancer
Authors Zhang M , Li X, Fan Z, Zhao J, Liu S, Zhang M, Li H, Goscinski MA, Fan H, Suo Z
Received 5 December 2017
Accepted for publication 6 February 2018
Published 28 May 2018 Volume 2018:11 Pages 3149—3157
DOI https://doi.org/10.2147/OTT.S158820
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ingrid Espinoza
Meng Zhang,1 Xiaoli Li,1,2 Zhirui Fan,1 Jing Zhao,1 Shuzheng Liu,3 Mingzhi Zhang,1 Huixiang Li,4 Mariusz Adam Goscinski,5 Huijie Fan,1 Zhenhe Suo1,2,6
1Department of Oncology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou University, Zhengzhou, Henan Province, China; 2Department of Pathology, The Norwegian Radium Hospital, Oslo University Hospital, University of Oslo, Oslo, Norway; 3Henan Office for Cancer Research and Control, Henan Cancer Hospital, Zhengzhou, Henan Province, China; 4Department of Pathology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan Province, China; 5Department of Surgery, The Norwegian Radium Hospital, Oslo University Hospital, University of Oslo, Oslo, Norway; 6Department of Pathology, Institute for Clinical Medicine, Faculty of Medicine, University of Oslo, Oslo, Norway
Background: Sushi repeat-containing protein X-linked 2 (SRPX2) is overexpressed in a variety of different tumor tissues and correlated with poor prognosis in patients. Little research focuses on the role of SRPX2 expression in prostate cancer (PCa), and the clinicopathological significance of the protein expression in this tumor is relatively unknown. However, our previous transcriptome data from those cancer stem-like cells indicated the role of SRPX2 in PCa.
Materials and methods: In this study, RT-PCR and Western blotting were firstly used to examine the SRPX2 expression in three PCa cell lines including LNCaP, DU145, and PC3, and then SRPX2 protein expression was immunohistochemically investigated and statistically analyzed in a series of 106 paraffin-embedded PCa tissue specimens.
Results: Significantly lower levels of SRPX2 expression were verified in the LNCaP cells, compared with the expression in the aggressive DU145 and PC3 cells, in both mRNA and protein levels. Immunohistochemically, there were variable SRPX2 protein expressions in the clinical samples. Moreover, high levels of SRPX2 expression in the PCa tissues were significantly associated with Gleason score (P=0.008), lymph node metastasis (P=0.009), and distant metastasis (P=0.021). Furthermore, higher levels of SRPX2 expression in the PCa tissues were significantly associated with shorter overall survival (OS) (P<0.001).
Conclusion: Our results demonstrate that SRPX2 is highly expressed in aggressive PCa cells in vitro, and its protein expression in PCa is significantly associated with malignant clinical features and shorter OS, strongly indicating its prognostic value in prostate cancers.
Keywords: SRPX2, immunohistochemistry, prostate cancer
Introduction
Prostate cancer (PCa) has the highest incidence on a global scale for men, which is the third leading cause of cancer death among men. Between 1990 and 2013, the PCa incidence increased globally by 217%.1,2 The vast majority of patients in the early stages of PCa may achieve excellent long-term cure rates by radical prostatectomy, some form of radiation therapy, or androgen-deprivation therapy. However, 10%–20% of the patients develop castration-resistant prostate cancer within about 5 years of follow-up, and 30%–50% of men will experience biochemical prostate specific antigen (PSA) recurrence within 10 years after treatment.3,4 Now, it has been realized that novel powerful biomarkers of PCa for early assessment and effective therapeutic treatment are needed.
Sushi repeat-containing protein X-linked 2 (SRPX2) was found in pro-B acute leukemia for the first time in 1999. It was described as a downstream target gene for E2A-HLF fusion gene.5 The SRPX2 protein consists of 465 amino acid residues, including one DUF4174 domain, three sushi repeat domains, and one hyaline repeat (HYR) domain (GenBank NP-055282). Some of the initial studies have shown that SRPX2 plays a crucial role in the development of the cerebral language center. The mutations of SRPX2 were identified to lead to rolandic seizures, language or cognitive dysfunction, and mental retardation.6 In addition to expression in normal tissues in brain, heart, lung, and placenta, high levels of SRPX2 expression have been revealed in various tumor tissues.7,8 As an extracellular matrix protein, SRPX2 has been shown to be involved in proliferation, migration, adhesion, and invasion of tumor cells through different signaling pathways and promote tumor progress.
In our previous studies, human PCa cell line DU145 was treated with either mtDNA replication reagent ethidium bromide or cell cycle inhibitor flavopiridol for years, and those cells that survived such a long-term treatment were functionally inhibited by mitochondria, and the cells were revealed with typical Warburg and cancer stem cell features. Comparative transcriptomic data from these cells revealed significantly higher levels of SRPX2 compared with their parental control cells,9,10 indicating a possible role of SRPX2 in PCa. To explore the clinical significance of SRPX2 in this study, we first examined its expression in different PCa cell lines and discovered low level of SRPX2 expression in the LNCaP cell line, compared with its expression in other aggressive PCa cell lines DU145 and PC3. Based on the above findings, we further immunohistochemically examined the expression of SRPX2 in a series of 106 PCa tumors and 10 noncancerous prostate tissue samples. The correlations between SRPX2 protein expression and conventional clinicopathological features were statistically explored, and its survival correlation was analyzed with the Kaplan–Meier method.
Materials and methods
Ethics statement
This study was approved by the Institutional Ethics Review Board of Zhengzhou University, Zhengzhou, China. All involved patients agreed to participate in this study and submitted written informed consent at the First Affiliated Hospital of Zhengzhou University.
Cell lines
Three PCa cell lines LNCaP, DU145, and PC3 were purchased from American Type Culture Collection (Manassas, VA, USA) and sustained in our laboratory for the research. The three cell lines were derived from lymph node metastases, brain metastases, and bone metastases in patients with PCa, respectively. Cells were routinely cultured in RPMI-1640 medium (11835-063; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, 10500-064; Thermo Fisher Scientific), 100 U/mL of penicillin, and 100 μg/mL of streptomycin (15140122; Thermo Fisher Scientific) at 37°C in a 5% CO2 atmosphere.
RT-PCR
Cells in 80% confluence were washed with cold PBS before 1.0 mL Trizol reagent was added to extract total RNA. Then the RNA was precipitated by isopropanol, washed with 75% ethanol, and dissolved using 20 μL 0.1% DEPC. The RNA sample was then reverse transcribed into cDNA by means of the RevertAid First Strand cDNA Synthesis Kit (#K1622; Thermo Fisher Scientific) according to the manufacturer’s instruction. The synthesized cDNA was used for PCR by using FastStart Universal SYBR Green Master (Rox) (04 913 914 001; Hoffman-La Roche Ltd., Basel, Switzerland) on the StepOnePlus Real-Time PCR System (Thermo Fisher Scientific). The primers used for PCR were as follows: forward primer GCTCTCTTTCTGCTGTTCTTCCT; reverse primer TGTATAACACCATCGGGGGAC. The analysis of all samples was repeated at least three times, and the relative quantitative results were analyzed by 2−ΔΔCt values.
Western blotting
All the cells at 80% confluence were harvested by using cell scraper, and the cells were washed twice with PBS at 4°C. They were then lysed with RIPA buffer (89900; Thermo Fisher Scientific) containing 100 mM PMSF (1862209; Thermo Fisher Scientific) on ice for 30 minutes. Cell lysates were centrifuged at 12,000 g for 10 minutes at 4°C, and then total protein concentration was investigated using BCA reagent (Beyotime, Beijing, China). Aliquots of 20 μg proteins mixed with sodium dodecyl sulfate (SDS)-loading buffer were boiled for 10 minutes. The protein samples were subjected to 10% SDS-polyacrylamide gel electrophoresis before electro-transferred onto high-quality polyvinylidene difluoride membranes in a Trans-Blotting apparatus (Bio-Rad Laboratories Inc., Hercules, CA, USA). After blocking with 5% BSA blocking buffer for 1 hour at room temperature, the membranes were incubated at 4°C overnight with mouse antihuman SRPX2 monoclonal (66266-1-Ig; Proteintech, Wuhan, China). The membranes were washed three times with TBST (TBS with 0.1% Tween) and incubated with horseradish peroxidase (HRP)-conjugated secondary antibody diluted 3,000 times with TBST at room temperature for 1 hour. Finally, immunoreactive bands were visualized with an enhanced chemiluminescence detection reagent (Beyotime). The protein bands were normalized to ACTIN and quantified by using Image Lab 2.0 Software (Bio-Rad Laboratories Inc.).
Patients
One hundred and six cases of formalin-fixed and paraffin-embedded PCa specimens were randomly included in this study. All the patients were diagnosed and surgically treated at the First Affiliated Hospital of Zhengzhou University between December 2005 and August 2017. All patients treated with chemotherapy, hormone therapy, or radiotherapy before surgery were excluded from this study. All patients were followed up from the date of confirmed diagnosis until death or August 2017. The clinicopathological information of the 106 PCa patients is summarized in Table 1. The ages of all patients ranged from 51 to 92 years with a median age of 70.5 years. In accordance with the Gleason score system, there were 34 low- (4–6), 43 moderate- (7), and 29 (8–10) high-grade tumors. The TNM staging following the Would Health Organization (WHO) standard showed 56 stage II patients and 50 stage III and IV patients. All involved specimens were reviewed and diagnosed by at least two pathologists from the First Affiliated Hospital of Zhengzhou University.
  | Table 1 Clinical and pathological characteristics for 106 patients with malignant prostate cancer |
Immunohistochemistry (IHC)
The SPlink Detection Kits (Biotin-Streptavidin HRP Detection Systems, SP9000, ZSGB-BIO, Beijing, China), were applied for IHC detection of SRPX2 according to the manufacturer’s instruction. Paraffin-embedded PCa tissues were cut into 3 μm sections, dried at 60°C for 2 hours and stored at 4°C. Before IHC, the paraffin sections were deparaffinized and then microwaved in citrate buffer (pH 6.0) for 15 minutes for antigen retrieval. The slides were then allowed to recover to room temperature and washed with PBS. To restrict the activity of endogenous peroxidase and avoid nonspecific staining, the slides were blocked with H2O2 (3%) and goat serum for 15 minutes at room temperature. They were then incubated with SRPX2 antibody (1:400, ab91584, Abcam, Shanghai, China) at 4°C overnight, followed by incubation with biotin-labeled goat anti-mouse/rabbit immunoglobulin G (IgG) polymer for 15 minutes and HRP for 30 minutes at room temperature. Whereafter, all slides were stained with 3,3′-diaminobenzidine tetrahydrochloride for 3–5 minutes and counterstained with hematoxylin, dehydrated, and mounted. The human lung tissue, always positive for SRPX2 staining, was used as positive control in our study. For negative controls, the primary antibody was replaced with nonimmune rabbit IgG with the same concentration in this study.
Immunohistochemical scoring
The immunostaining of SRPX2 was evaluated by two pathologists from the Department of Pathology of the First Affiliated Hospital of Zhengzhou University with consensus. The evaluators were blinded to the clinical data of patients. The level of SRPX2 expression in PCa samples was assessed by semiquantitative evaluation of immunohistochemical staining, including the assessment of the staining intensity and staining extent of the samples. For the scoring of staining extent, samples without stained cells were rated as 0, those with <25% stained cells as 1, those with 25%–50% stained cells as 2, and those with >50% stained cells were rated as 3. The staining intensity was counted as 0, 1, 2, and 3 when the outcomes of staining revealed no positive cells, weak staining, moderate staining, and strong staining, respectively. The results of immunohistochemical staining scores are shown in Table 2. The samples were grouped as negative for a score of 0 (−), moderately positive for a score of 1–4 (+), and strongly positive for a score of 5–6 (++) according to the final score (the sum of the extent and intensity score).
Statistical analysis
Statistical analysis was performed using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). Data such as SRPX2 expression in mRNA or protein level were analyzed using the Student’s t-test. Chi-square tests (Pearson’s and linear-by-linear, as appropriate) or Fisher’s exact probabilities were used for assessment of the associations between SRPX2 expression and clinicopathological characteristics. Survival analysis was performed using the Kaplan–Meier method, and groups were compared with two-sided log-rank tests. Univariate and multivariate survival analyses were accomplished using Cox proportional regression hazard model to identify the significant factors that were involved in prognosis. For all the analyses, P<0.05 was considered to be significant.
Results
SRPX2 expression in PCa cell lines
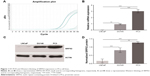
Variable SRPX2 expression could be detected in the human PCa cell lines LNCaP, DU145, and PC3. However, compared to those in the aggressive DU145 and PC3 cell lines, significantly lower levels of SRPX2 expression were verified in the LNCaP cell line with both RT-PCR in mRNA and Western blotting in protein levels (Figure 1).
SRPX2 expression in PCa tissues
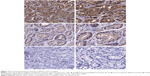
There were 106 cases of PCa specimens and 10 noncancerous prostate tissues adjacent to tumor that were detected by immunohistochemical staining to clarify the SRPX2 expression in this study. It turned out that SRPX2 protein was significantly overexpressed in PCa tissues. The typical cytoplasmic staining of SRPX2 protein in PCa cells is shown in Figure 2. Of the total 106 samples, 69 (65.10%) cases were strongly positive, 30 (28.30%) were moderately positive, while only seven (6.60%) were negative for SRPX2 expression. However, the noncancerous prostate tissues were all negative for SRPX2 expression.
Correlations between SRPX2 expression and clinicopathological features
As summarized in Table 3, the correlations of SRPX2 protein expression with the clinicopatological features were analyzed by chi-square test or Fisher’s exact test. Results indicated that SRPX2 protein expression was positively correlated with Gleason score (P=0.008). In addition, it was also found that the SRPX2 protein expression in PCa samples was positively associated with lymph node metastasis (P=0.009) and distant metastasis (P=0.021). No significant association between age (P=0.952), PSA (P=0.124), and Union for International Cancer Control (UICC) stage (P=0.117) of patients, and SRPX2 protein expression was discovered in this study.
  | Table 3 The associations of SRPX2 expression levels and the clinicopathological characteristics in prostate cancer |
Increased SRPX2 expression is significantly associated with unfavorable survival in PCa
The overall survival (OS) rate of the 106 PCa patients was 37.7%. A total of 66 deaths were observed in the 106 patients during the follow-up period. The follow-up time ranged from 2 to 147 months with a median follow-up time of 58 months. Of all the patients, seven cases of negative SRPX2 expression were observed, while 30 and 69 cases were moderately and strongly positive for the protein expression. As shown in Figure 3, Kaplan–Meier survival curve and log-rank test proved that high SRPX2 protein expression in PCa was significantly correlated with poor OS of patients (P<0.001). The survival rates of patients with SRPX2 negative, moderate, and strong expressions were 71.42%, 66.67%, and 21.74%, respectively.
SRPX2 expression in PCa is an independent risk factor for OS
Univariate and multivariate analyses were performed using Cox proportional hazards regression method on the basis of the above clinicopathological parameters and the expression of SRPX2 in tumor cells. As shown in Table 4, Gleason score (hazard ratio [HR] =1.314, 95% confidence interval [CI]: 1.043–1.657, P=0.021), UICC stage (HR =1.425, 95% CI: 1.021–1.990, P=0.037), and SRPX2 expression (HR =1.461, 95% CI: 1.118–1.909, P=0.006) were identified as independent risk factors for shorter OS in the PCa patients.
  | Table 4 Univariate and multivariate analysis for overall survival using Cox relative risk |
Discussion
PCa is a highly heterogeneous disease. The main clinical treatments of PCa include radical prostatectomy, radiation therapy, chemotherapy, and androgen ablation. In recent years, the diagnosis and therapy of this disease have been greatly improved, but challenges remain in regard to relapse and castration resistance after receiving treatment. On account of the Western lifestyle and the changing in dietary pattern at present, the incidence of PCa in China has increased steadily.11 The prognostic evaluation of patients with PCa is still largely dependent on Gleason score, level of PSA, surgical margin status, lymph node metastases, and TNM staging.12–14 Specific molecular markers as prognostic indicators for PCa are still missing.
SRPX2 is a novel chondroitin sulfate proteoglycan,7 which has been shown to participate in a series of biological processes of tumor cells. SRPX2 enhances the migration and adhesion of gastric cancer cells directly or indirectly by increasing the phosphorylation levels of focal adhesion kinase (FAK).15 Similarly, SRPX2 promotes the migration and invasion of pancreatic ductal adenocarcinoma cells through FAK pathway.16 In addition, studies have shown that silencing SRPX2 in colon cancer HCT116 cells remarkably impairs the proliferation, adhesion, migration, and invasion of cancer cells. On the contrary, elevated expression of SRPX2 in SW480 cells promotes cell migration and invasion. Besides, downregulation of SRPX2 protein resulted in a significant downregulation of β-catenin, MMP2/9, suggesting that SRPX2 may promote the invasion of colorectal cancer by regulating MMP2/9 expression through Wnt/β-catenin signaling pathway.17
Several previous studies have reported that SRPX2 is highly expressed in various malignant tumor tissues, such as gastric cancer,18 colorectal cancer,17 pancreatic cancer,16 and glioblastoma.19 Overexpression of SRPX2 is significantly associated with unfavorable outcomes in these patients. In agreement with the above studies, we first noticed the upregulation of SRXP2 in the PCa stem-like cells in our laboratory.9,10 Further study of the PCa cell lines LNCaP, DU145, and PC3 verified significantly higher levels of SRPX2 expression in those aggressive cell lines DU145 and PC3, while the biological rather mild cell line LNCaP expressed significantly lower levels of SRPX2, in both mRNA and protein levels.
Therefore, we further examined the expression of SRPX2 in a series of 106 cases of formalin-fixed and paraffin-embedded PCa tissues in the present study. Immunohistochemical staining showed that SRPX2 was highly expressed in PCa with a positive rate of 93.40%. We next investigated the relationship between SRPX2 expression and clinicopathological features in PCa. In the present study, high levels of SRPX2 protein expression in the 106 PCa samples were significantly correlated with the high Gleason score, lymph node metastasis, and distant metastasis. Accordingly, we considered that the overexpression of SRPX2 might contribute to the development and progression of PCa. Indeed, further survival analyses revealed that high levels of SRPX2 protein expression in PCa were significantly associated with shorter OS, indicating a poor prognostic factor of SRPX2 in PCa. Univariate and multivariate Cox proportional hazards regression analyses showed that SRPX2 expression was a significantly independent predictor of prognosis in PCa patients. Thus, our results merit extensive SRPX2 studies in terms of its function, predictive and prognostic values in PCa.
As mentioned above, SRPX2 plays an important role in the adhesion, migration, and invasion of various tumor cells. Existing in a variety of proteins with different functions, the CCP domains of SRPX2 protein influence the cell adhesion by mediating specific protein–protein and protein–carbohydrate interactions.8,20 The HYR domain plays a role in cell adhesion process.21 Schwanzer-Pfeiffer et al have suggested that SRPX2 may influence cell adhesion by regulating ICAM1, E-selectin shedding and the expression of membrane bound adhesion molecules.22 The results of phylogenetic analysis suggested that SRPX2 and selectin families are highly homologous and belong to the same gene family emerged during vertebrate evolution.23 It has been reported that selectin promotes the interaction between tumor cells and endothelial cells, which leads to tumor progression and metastasis.24,25 Besides, selectin is conducive to promoting angiogenesis.26,27 As an extracellular matrix protein,28 SRPX2 is involved in the regulation of extracellular matrix protein remodeling through its proteolytic activity. Royer-Zemmour et al demonstrated that SRPX2 is a ligand for urokinase-type plasminogen activator receptor and interacted with CTSB and ADAMTS4,29 both of which belong to the important components of extracellular proteolysis machinery. CTSB is an activator of uPA. Moreover, it has been found that SRPX2 is overexpressed in HUVEC.15 SRPX2 interacts with uPAR in endothelial remodeling during the early phases of angiogenesis.24 These studies suggest that SRPX2 protein is conducive to promote angiogenesis. Recently, Raza and Jaiswal constructed a PCa-specific gene regulatory network using novel simple statistics based on an approach extracting differentially expressed genes and found regulatory relationships among the genes in PCa. The results indicate a potential link between EMP1, CSTF1, and SRPX2 genes in PCa and predict the role of SRPX2 as target for PCa.30
Conclusion
The current study reveals significantly higher levels of SRPX2 expression in aggressive PCa cell lines, and overexpression of SRPX2 protein in PCa predicts unfavorable clinicopathological features and poor outcome of patients. Consequently, it may be used as a novel prognostic marker and a potential target for PCa treatment. It should be noted that the present study has explored only a limited number of PCa samples, and the crucial roles of SRPX2 in PCa and the relevant molecular mechanisms deserve to be further elucidated.
Acknowledgment
This study was financially supported by The Radium Hospital Legat and a grant from National Natural Science Foundation of China (81272824).
Disclosure
The authors report no conflicts of interest in this work.
References
Global Burden of Disease Cancer Collaboration. The Global Burden of Cancer 2013. JAMA Oncol. 2015;1(4):505–527. | ||
Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. | ||
Kirby M, Hirst C, Crawford ED. Characterising the castration-resistant prostate cancer population: a systematic review. Int J Clin Pract. 2011;65(11):1180–1192. | ||
Pound CR, Brawer MK, Partin AW. Evaluation and treatment of men with biochemical prostate-specific antigen recurrence following definitive therapy for clinically localized prostate cancer. Rev Urol. 2001;3(2):72–84. | ||
Kurosawa H, Goi K, Inukai T, et al. Two candidate downstream target genes for E2A-HLF. Blood. 1999;93(1):321–332. | ||
Roll P, Rudolf G, Pereira S, et al. SRPX2 mutations in disorders of language cortex and cognition. Hum Mol Genet. 2006;15(7):1195–1207. | ||
Tanaka K, Arao T, Tamura D, et al. SRPX2 is a novel chondroitin sulfate proteoglycan that is overexpressed in gastrointestinal cancer. PLoS One. 2012;7(1):e27922. | ||
O’Leary JM, Bromek K, Black GM, et al. Backbone dynamics of complement control protein (CCP) modules reveals mobility in binding surfaces. Protein Sci. 2004;13(5):1238–1250. | ||
Li XR, Zhong YL, Lu J, et al. MtDNA depleted PC3 cells exhibit Warburg effect and cancer stem cell features. Oncotarget. 2016;7(26):40297–40313. | ||
Li XR, Lu J, Kan QC, et al. Metabolic reprogramming is associated with flavopiridol resistance in prostate cancer DU145 cells. Sci Rep. 2017;7(1):5081. | ||
Zhu Y, Wang HK, Qu YY, Ye DW. Prostate cancer in East Asia: evolving trend over the last decade. Asian J Androl. 2015;17(1):48–57. | ||
Kattan MW, Vickers AJ, Yu C, et al. Preoperative and postoperative nomograms incorporating surgeon experience for clinically localized prostate cancer. Cancer. 2009;115(5):1005–1010. | ||
Capitanio U, Briganti A, Gallina A, et al. Predictive models before and after radical prostatectomy. Prostate. 2010;70(12):1371–1378. | ||
Han M, Partin AW, Zahurak M, Piantadosi S, Epstein JI, Walsh PC. Biochemical (prostate specific antigen) recurrence probability following radical prostatectomy for clinically localized prostate cancer. J Urol. 2003;169(2):517–523. | ||
Tanaka K, Arao T, Maegawa M, et al. SRPX2 is overexpressed in gastric cancer and promotes cellular migration and adhesion. Int J Cancer. 2009;124(5):1072–1080. | ||
Gao Z, Zhang J, Bi M, et al. SRPX2 promotes cell migration and invasion via FAK dependent pathway in pancreatic cancer. Int J Clin Exp Pathol. 2015;8(5):4791–4798. | ||
Liu KL, Wu J, Zhou Y, Fan JH. Increased Sushi repeat-containing protein X-linked 2 is associated with progression of colorectal cancer. Med Oncol. 2015;32(4):99. | ||
Yamada T, Oshima T, Yoshihara K, et al. Impact of overexpression of Sushi repeat-containing protein X-linked 2 gene on outcomes of gastric cancer. J Surg Oncol. 2014;109(8):836–840. | ||
Tang H, Zhao J, Zhang L, Zhao J, Zhuang Y, Liang P. SRPX2 enhances the epithelial-mesenchymal transition and temozolomide resistance in glioblastoma cells. Cell Mol Neurobiol. 2016;36(7):1067–1076. | ||
Soares DC, Gerloff DL, Syme NR, Coulson AF, Parkinson J, Barlow PN. Large-scale modelling as a route to multiple surface comparisons of the CCP module family. Protein Eng Des Sel. 2005;18(8):379–388. | ||
Callebaut I, Gilgès D, Vigon I, Mornon JP. HYR, an extracellular module involved in cellular adhesion and related to the immunoglobulin-like fold. Protein Sci. 2000;9(7):1382–1390. | ||
Schwanzer-Pfeiffer D, Roßmanith E, Schildberger A, Falkenhagen D. Characterization of SVEP1, KIAA, and SRPX2 in an in vitro cell culture model of endotoxemia. Cell Immunol. 2010;263(1):65–70. | ||
Royer B, Soares DC, Barlow PN, et al. Molecular evolution of the human SRPX2 gene that causes brain disorders of the Rolandic and Sylvian speech areas. BMC Genet. 2007;8:72. | ||
Miljkovic-Licina M, Hammel P, Garrido-Urbani S, Bradfield PF, Szepetowski P, Imhof BA. Sushi repeat protein X-linked 2, a novel mediator of angiogenesis. FASEB J. 2009;23(12):4105–4116. | ||
Barthel SR, Gavino JD, Descheny L, Dimitroff CJ. Targeting selectins and selectin ligands in inflammation and cancer. Expert Opin Ther Targets. 2007;11(11):1473–1491. | ||
Vernes SC, Oliver PL, Spiteri E, et al. Foxp2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS Genet. 2011;7(7):e1002145. | ||
Egami K, Murohara T, Aoki M, Matsuishi T. Ischemia-induced angiogenesis: role of inflammatory response mediated by P-selectin. J Leukoc Biol. 2006;79(5):971–976. | ||
Wilson R, Norris EL, Brachvogel B, et al. Changes in the chondrocyte and extracellular matrix proteome during post-natal mouse cartilage development. Mol Cell Proteomics. 2012;11(1):M111.014159. | ||
Royer-Zemmour B, Ponsole-Lenfant M, Gara H, et al. Epileptic and developmental disorders of the speech cortex: ligand/receptor interaction of wild-type and mutant SRPX2 with the plasminogen activator receptor uPAR. Hum Mol Genet. 2008;17(23):3617–3630. | ||
Raza K, Jaiswal R. Reconstruction and analysis of cancer-specific gene regulatory networks from gene expression profiles. Int J Bioinform Biosci. 2013;3(2):25–34. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.