Back to Journals » Infection and Drug Resistance » Volume 13
High-Level Resistance of Toxigenic Clostridioides difficile Genotype to Macrolide-Lincosamide- Streptogramin B in Community Acquired Patients in Eastern China
Authors Zhao L, Luo Y, Bian Q, Wang L, Ye J, Song X, Jiang J, Tang YW , Wang X, Jin D
Received 16 November 2019
Accepted for publication 26 December 2019
Published 17 January 2020 Volume 2020:13 Pages 171—181
DOI https://doi.org/10.2147/IDR.S238916
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Longyou Zhao, 1,* Yun Luo, 2,* Qiao Bian, 3, 4 Liqian Wang, 5 Julian Ye, 6 Xiaojun Song, 4, 7 Jianmin Jiang, 6, 8 Yi-Wei Tang, 9–11 Xianjun Wang, 5 Dazhi Jin 4, 7
1Lishui Second People’s Hospital, Lishui, Zhejiang, People’s Republic of China; 2School of Biotechnology and Biomolecular Sciences, University of New South Wales, Sydney, NSW, Australia; 3School of Medicine, Ningbo University, Ningbo, Zhejiang, People’s Republic of China; 4Centre of Laboratory Medicine, Zhejiang Provincial People Hospital, People’s Hospital of Hangzhou Medical College, Hangzhou, Zhejiang, People’s Republic of China; 5Department of Laboratory Medicine, Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, People’s Republic of China; 6Department of Microbiology, Zhejiang Provincial Center for Disease Control and Prevention, Hangzhou, Zhejiang, People’s Republic of China; 7School of Laboratory Medicine, Hangzhou Medical College, Hangzhou, Zhejiang, People’s Republic of China; 8Key Laboratory of Vaccine, Prevention and Control of Infectious Disease of Zhejiang Province, Hangzhou, Zhejiang, People’s Republic of China; 9Department of Laboratory Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA; 10Department of Pathology and Laboratory Medicine, Weill Medical College of Cornell University, New York, NY, USA; 11Cepheid, Danaher Diagnostic Platform, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Dazhi Jin
Centre of Laboratory Medicine, Zhejiang Provincial People Hospital, People’s Hospital of Hangzhou Medical College, No. 158 Shangtang Road, Hangzhou, Zhejiang 310014, People’s Republic of China
Tel/Fax +86-571-87692692
Email [email protected]
Xianjun Wang Tel/Fax +86-571-56007158
Email [email protected]
Background: Clostridioides difficile resistant to macrolide-lincosamide-streptogramin B (MLSB) has not been reported in China.
Methods: In a cross-sectional study in two tertiary hospitals, C. difficile isolates from stool specimens from community-onset, hospital-associated diarrheal patients were analyzed for toxin genes, genotype, and antibiotic resistance, and the patients’ clinical charts were reviewed.
Results: A total of 190 (15.2%) isolates (102 A+B+ and 88 A−B+) from 1250 community acquired (CA) patients were recovered and all were susceptible to vancomycin and metronidazole. High-level resistance (minimum inhibitory concentration > 128 mg/L) to erythromycin and clindamycin was recorded in 77.9% and 88.4% of the tested isolates, respectively. Furthermore, 89.3% (159/178) of the isolates resistant to MLSB carried the erythromycin resistance methylase gene (ermB). The statistically significant factors associated with C. difficile infection (CDI) induced by A−B+ isolates with MLSB resistance included a severity score of > 2 (odds ratio [95% confidence interval], 7.43 [2.31– 23.87]) and platelet count (cells × 10 9 cells/L) < 100 [5.19 (1.58– 17.04)]. The proportion of A−B+ increased with enhanced CDI severity (x 2 = 21.62, P < 0.001), which was significantly higher than that of ermB-positive A+B+ in severity score of 4 (x 2 = 8.61, P = 0.003). The average severity score of ermB-positive isolates was significantly higher than that of ermB-negative isolates in A−B+ (Z = − 2.41, P = 0.016).
Conclusion: The ermB-positive A−B+ C. difficile with MLSB resistance is described for the first time as a potential epidemic clone inducing severe CDI in CA diarrheal patients in Eastern China.
Keywords: Clostridioides difficile, molecular characteristic, macrolide-lincosamide-streptogramin B resistance
Introduction
Clostridioides difficile, an anaerobic, gram-positive, spore-forming bacterium, causes severe infectious colitis, toxic megacolon, and sepsis, which are life threatening, and it is also responsible for antibiotic-associated diarrhea exhibiting asymptomatic carriage or mild manifestations.1 Clostridioides difficile infection (CDI) affects approximately more than 300,000 hospitalized patients each year2–6 and has become the leading cause of hospital-acquired diarrhea worldwide.7,8
Exposure to antibiotics is considered the most important modifiable risk factor for the development of CDI.9 Antibiotic resistance of C. difficile also plays a key role in driving the emergence of new strain types. C.difficile PCR ribotype 027 has a high correlation with fluoroquinolone resistance, leading to its widespread distribution in North America and Europe.10 Metronidazole and vancomycin remain the first-line therapeutics for CDI despite the emergence of sporadic metronidazole- and vancomycin-resistant C. difficile.10 Resistance to macrolide-lincosamide-streptogramin B (MLSB) is considered an important factor not only driving the persistence and transmission of C. difficile but also increasing the risk for CDI in patients.11
Recently, the rate of MLSB resistance has been significantly increasing worldwide. The antibiotic resistance profile of C. difficile in 2012–2015 revealed that the rate of erythromycin (ERY) resistance ranged from 13% to 100% and that of clindamycin (CLI) resistance ranged from 8.3% to 100%.10 The emergence of CLI-resistant 027 has been found to be a new strain type increasing the risk of CDI and accelerating its spread in Europe.11,12 In 2001, a CLI-resistant, toxin A-negative, and toxin B-positive (A−B+) C. difficile clone with the ermB gene has been reported be associated with a nosocomial outbreak in the Netherlands. This clone was also a potential predictor of enhanced CDI in Poland.13 A systematic review and meta-analysis from 2010 to 2016 in China showed that the rate of ERY and CLI resistance was 80.2% and 81.7%, respectively.14 A cross-sectional study in Eastern China in 2012–2015 revealed that resistance to ERY and CLI was 64.7% and 62.5%, respectively, with significantly different distribution in different genotypes.15
MLSB-type antibiotic resistance is due to post-transcriptional methylation of 23S ribosomal rRNA.16 The resistance of C. difficile to ERY and CLI has been described to be transferred with no involvement of plasmid DNA. The resistance of C. difficile to ERY and CLI is encoded by the erythromycin resistance methylase gene (ermB) located on the mobilizable non-conjugative transposon Tn 5398.16,17 However, ermB genes have not been identified in all clinical C. difficile isolates expressing high resistance to ERY and CLI, which might be conferred by other mechanisms. Moreover, the data on MLSB resistance of C. difficile in China are limited. The rate of MLSB resistance and the correlation between genotypes and MLSB resistance and between C. difficile carrying ermB genes and virulence properties are still unclear.
The present cross-sectional study in community-onset, hospital-associated diarrheal patients from two hospitals in Eastern China was conducted to reveal the correlations among toxin genes, MLSB resistance, and ermB genes. We also analyzed the difference in CDI severity induced by different genotypes with MLSB resistance.
Materials and Methods
Sample Collection
Clinical samples were consecutively collected from December 2015 to April 2016 from two hospitals, the Affiliated Hangzhou First People’s Hospital (HFPH), Zhejiang University school of Medicine and the Lishui Second People’s Hospital (LSPH), which are located in Zhejiang Province, China. The HFPH is a general hospital with 2613 beds in 86 units. The LSPH is a general hospital with 500 beds. Clinical stool specimens from patients with suspected CDI were transported to the Zhejiang Provincial Center for Disease Control and Prevention (ZJCDC) within 72 h for further testing. This study was approved by the Institutional Review Board of the ZJCDC, LSPH, and HFPH.
According to the guidelines of the Society for Healthcare Epidemiology of America and the Infectious Diseases Society of America (SHEA/IDSA),9 inclusion criteria were presented as follows. We collected stool specimens from patients who were admitted within less than 48 h with diarrhea having loose, watery, or unformed stools more than three times within 24 h. Exclusion criteria were detection of other diarrhea-causing pathogens in the stool specimens, neonates or infants under 12 months of age, and patients with diarrhea who have been admitted over 48 h. Duplicated stool specimens from the same patients were removed. A standardized questionnaire including basic information and clinical data was prepared for each patient. All clinical parameters as part of routine care were recorded with biochemical and immunological examinations, and the cutoff values of stool-red blood cells (S-RBCs), stool-white blood cells (S-WBCs), occult blood in stool (OB), C-reactive protein (CRP), white-blood cells (WBCs), neutrophils, lymphocytes, monocytes, eosinophils, basophils, hemoglobin (Hb), platelets (PLT), creatinine, glucose, and kalium were obtained from the clinical standards.18,19 CDI was diagnosed, clinical cases were classified into category of CDI based on the clinical and laboratory results, and hospital acquired and community acquired (CA) CDI as reported by the SHEA/IDSA.9 Six categories of CDI severity were evaluated mainly based on clinical manifestations of the patients and the laboratory results. Each patient was classified into one of six severity scores, which was 1 (no clinical CDI), 2 (mild), 3 (mild to moderate), 4 (moderate), 5 (moderate to severe), and 6 (severe) as reported.15,20,21
Clostridioides difficile Culture
Stool specimens were treated with alcohol, and then the mixture was inoculated onto cycloserine-cefoxitin fructose agar with selective supplement (Oxoid, Basingstoke, UK) as previously reported.15 The isolates were identified by morphological characteristics of flat, yellow, ground-glass appearance, and special odor on the CCFA.22 C. difficile was further confirmed using the latex agglutination test (Oxoid, Ltd., Basingstoke, UK). All isolates were stored at −70°C in the brain–heart infusion broth with 10% glycerol until subsequent analyses.
Detection of C. difficile Toxin and ermB Genes
Genomic DNA of C. difficile was extracted using the QIAamp DNA Blood Mini Kit (Valencia, CA, USA), according to the manufacturer’s instructions. The toxin genes tcdA and tcdB and the housekeeping gene tpi were detected using a conventional polymerase chain reaction (PCR) assays with the primer sequences reported previously.15 The ermB gene was detected by the conventional PCR assay as previously reported.13 After PCR amplification, the PCR products were analyzed by agarose gel electrophoresis. The tcdA-F and tcdA-R primers were used to detect the tcdA gene, yielding a 369 bp amplicon for tcdA-positive strains or a 110 bp amplicon for tcdA-negative strains. The PCR product of ermB gene was 688 bp. Two standard C. difficile strains (ATCC 43255 and 700057) obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) were used as the positive and negative controls for both tcdA and tcdB, respectively. The blank, positive, and negative controls were examined in each test simultaneously.
Multilocus Sequence Typing
Six reference C. difficile strains, namely, ATCC 43255, 43598, BAA-1870, BAA-1803, BAA-1801, and 700057, were used as the controls. All C. difficile isolates were genotyped by multilocus sequence typing (MLST) as described previously.23 In brief, seven loci, namely, adk, atpA, dxr, glyA, recA, sodA, and tpi, widely distributed on the chromosome, were amplified by the PCR. The PCR products were sequenced using the 3730 XL DNA Analyzer (Applied Biosystems). All the sequences according to the seven loci were input into the C. difficile MLST database (http://pubmlst.org/cdifficile) for determining the sequence types (STs). A minimal spanning tree was constructed using Bionumerics software version 5.1 (Applied Math, Austin, TX, USA).
Antibiotic Susceptibility Testing
The isolates of C. difficile were tested for resistance to metronidazole, vancomycin, clindamycin, and erythromycin by agar dilution as previously reported.24 Bacteroides fragilis (ATCC 25285) and C. difficile (ATCC 700057) were included in each run as the controls. The minimal inhibitory concentration (MIC) results were interpreted following the guidelines of the Clinical and Laboratory Standards Institute (CLSI) published in 2017 for metronidazole and clindamycin.24 The breakpoints for vancomycin and erythromycin were determined according to a previous study.25
Data Analysis
The data were analyzed using Statistical Package for Social Sciences (SPSS, Chicago, IL, USA) version 19.0 and Epi Info version 3.5.1. The difference in CDI severity score among different genotypes was analyzed using the nonparametric test. Logistic regression analysis was used to identify independent risk factors. The odds ratio (OR), 95% confidence interval (CI), and P-values were calculated to assess the differences in clinical characteristics between A+B+ and A−B+ isolates resistant to CRY and CLI. The results with P-values < 0.05 were considered statistically significant.
Results
Clinical Information of Diarrheal Patients from Two Hospitals
A total of 1250 patients with diarrhea at the HFPH (n = 1053) and LSPH (n = 197) were enrolled in this cross-sectional study conducted over five months. One stool specimen was collected from each patient, with no duplicated specimens, when the patients were admitted within less than 24 h. We recovered a total of 190 (15.2%) isolates (HFPH, n = 141; LSPH, n = 49) with the tpi gene from these stool specimens. All the isolates were toxigenic C. difficile carrying either or both tcdA and tcdB; no nontoxigenic isolates were found (Figure 1). All the clinical information, including basic information and immune-biochemical data, was compared between the patients from the HFPH and LSPH (Table 1). All 190 patients had CA CDI. The CDI rate in patients from the HFPH was 13.4%, which was significantly lower than that in patients from the LSPH (24.9%) (χ2 = 20.40, P < 0.0001). The patients enrolled in this study were mainly from the wards of infectious diseases, psychiatry and neurology, and blood and cardiology in the LSPH; however, the wards of gastroenterology, infectious disease, and pediatrics were the main sources in the HFPH, which resulted in the difference in ward distribution between the LSPH and HFPH (χ2 = 429.22, P < 0.0001).
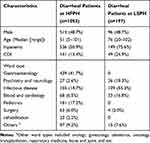 |
Table 1 Clinical Information of Diarrheal Patients Involved in This Study |
 |
Figure 1 Flow diagram of data collected during the study. |
CDI Severity
The clinical data of all the enrolled patients were assessed via blinded chart review by physicians as reported previously.15,20,21 No case was categorized under the CDI severity scores of 1, 5, and 6. Furthermore, 69.5% of CDI was categorized under the severity score 2 and 22.6% was under the severity score 3. Only 15 (7.9%) patients with CDI presented moderate clinical symptoms graded as CDI severity score 4. All the isolates were subjected to further analysis (Figure 1).
Genotype of C. difficile Isolates
Of the 190 toxigenic C. difficile, 102 (53.7%) isolates were positive for tcdA and tcdB (A+B+), in which ST3, ST35, and ST54 were the dominant genotypes. The 88 (46.3%) isolates were negative for tcdA but positive for tcdB (A−B+), among which three STs were ST37, ST39, and ST81. All the isolates were divided into 18 STs, indicating a high genetic diversity of C. difficile in these two hospitals, resulting in the difference in ST distribution between these hospitals (χ2 = 6.02, P = 0.049) (Table 2).
 |
Table 2 Molecular Characteristics of Toxigenic C. difficile Isolates in HFPH and LSPH |
A minimal spanning tree was constructed to reveal the allelic difference among isolates. As shown in Figure 2, two independent clusters (A and B) were obviously divided in the minimal spanning tree. The cluster A presented high diversity within 14 STs, and seven of them were completely resistant to MLSB. Half of the ST8 and ST233 isolates were susceptible to MLSB, and the other four STs had lower resistance rates, ranging from 6.5% to 33.3%. ST11 and all the A−B+ isolates, including ST37, ST39, and ST81, were included in cluster B with 100% MLSB resistance. The A−B+ isolates with MLSB resistance were closely related and had single or four different allelic genes in cluster B. The MLSB-susceptible isolates were distributed in cluster A in a disorderly manner, and no genetic relationship between MLSB-resistant and MLSB-susceptible C. difficile isolates was found. There were no differences on distribution of MLSB-resistant C. difficile isolates between two hospitals (χ2 = 3.13, P = 0.077), indicating that this clone was a common genotype in community associated CDI in Eastern China (Table 2).
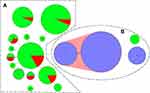 |
Figure 2 Relationship of toxigenic C. difficile isolates susceptible or resistant to MLSB by minimal spanning tree. |
Antibiotic Resistance and ermB of Isolates
The antibiotic-susceptibility pattern of 190 toxigenic C. difficile isolates is presented in Table 2. No isolates were resistant to metronidazole (MIC ≤ 8 mg/L) or vancomycin (MIC ≤ 2 mg/L). However, a high level of CLI resistance (MIC > 128 mg/L) was observed for all the isolates, including all the 49 isolates from the LSPH and the 119 isolates from the HFPH; the 13 isolates from the HFPH had intermediate susceptibility to CLI (MIC = 4 mg/L), and the remaining 9 were susceptible to CLI. The distribution of CLI resistant isolates were found statistically significant between HFPH and LSPH (χ2=8.65, P = 0.005). A total of 148 isolates, including 108 from the HFPH and 40 from the LSPH, were highly resistant to ERY (MIC >128 mg/L). The 88.2% of A+B+ (90/102) and 100% of A−B+ (88/88) C. difficile isolates were resistant to MLSB. The 12 ERY-susceptible isolates (6.3%, 12/190), including 4 isolates susceptible to CLI and 8 isolates intermediate-susceptible to CLI did not carry the ermB gene. The rate of carrying ermB in MLSB-resistant isolates was 89.3% (159/178). The 19 ermB-negative isolates (10.7%, 19/178) were simultaneously resistant to ERY and CLI, except one from the HFPH, which was resistant to CLI but susceptible to ERY.
Predictive Factors for Infection with MLSB-Resistant C. difficile
The basic information, CDI severity score, and immuno-biochemical data of the patients with CDI were analyzed and compared between A+B+ and A−B+ C. difficile isolates resistant to MLSB. A bivariate analysis between 90 A+B+ and 88 A−B+ isolates resistant to MLSB was performed, and the results are presented in Table 3. The number of patients infected by A−B+ C. difficile isolates resistant to MLSB was significantly more than the number of patients with A+B+ C. difficile isolates resistant to MLSB in CDI severity score of >2, WBC (cells × 109/mL) count of >10, and platelet (cells × 109/L) count of <100, respectively. A multivariate analysis including statistically significant parameters from the bivariate analysis was subsequently conducted. The percentages of patients infected by A−B+ C. difficile isolates resistant to MLSB was significantly more than those of patients with A+B+ C. difficile isolates resistant to MLSB in only two parameters, CDI severity score of >2 and platelet (cells × 109/L) count of <100, respectively. There were no significant differences among different ages ranging from >20 to >80 years of age. However, we notably found that CDI induced by MLSB-resistant A−B+ C. difficile was more severe than that by MLSB-resistant A+B+ C. difficile.
 |
Table 3 Differences of Clinical Characteristics Between A+B+ and A−B+ C. difficile with Resistance to MLSB |
Correlation Among CDI Severities, Toxin A/B Types, and ermB Gene
CDI severities were assessed as described above. The average ± standard deviation severity score of MLSB-resistant A+B+ and A−B+ isolates with or without the ermB gene was calculated (Table 4 and Table S1). The average CDI severity scores induced by ermB-positive isolates in A−B+ C. difficile were significantly higher than those induced by the ermB-positive isolates in A+B+ C. difficile isolates (Z = −4.68, P < 0.001) (Table S1) and ermB-negative isolates in A−B+ C. difficile isolates (Z = −2.41, P = 0.016) (Table 4), respectively. However, no statistically significant differences were found between the ermB-positive and ermB-negative A+B+ isolates (Z = −0.21, P = 0.836) (Table 4) and between A+B+ and A−B+ in the ermB-negative isolates (Z = −1.11, P = 0.267) (Table S1). The above results showed that ermB-positive A−B+ C. difficile isolates with MLSB-resistance led to more severe CDI cases than others.
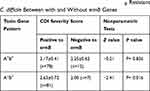 |
Table 4 Comparison of CDI Severity Scores in MLSB Resistant C. difficile Between with and Without ermB Genes |
Comparison Among CDI Severities, Genotypes, and ermB Gene in Isolates Resistant to MLSB
CDI severities, STs, and ermB gene were analyzed as follows in C. difficile isolates resistant to MLSB. In MLSB-resistant ermB-positive isolates, the proportion of A−B+ increased with severity score (x2 = 21.62, P < 0.001). The number of A−B+ isolates (91.7%, 11/12) was significantly more than the number of A+B+ isolates (8.3%, 1/12) with ermB in the category of CDI severity score of 4 (x2 = 8.61, P = 0.003) (Table 5). There were statistically significant differences in the CDI severity scores among different STs in the ermB-positive isolates (x2 = 36.77, P = 0.001) (Table 5). In patients with a CDI severity score of 4, a total of 9 ST37 genotype isolates (75.0%, 9/12), one of A−B+ C. difficile, was found, which was more frequently than other STs, and the number of ST37 increased with the CDI severity score (x2 = 26.38, P < 0.001) (Table 5). In ermB-negative isolates, only one patient had severe CDI and was categorized under the CDI severity score of 4. Most of the ermB-negative isolates had mild CDI with a CDI severity score of 2. Obviously, the ermB-positive A−B+ C. difficile isolates with MLSB-resistance, especially ST37, were the main drivers to induce increased CDI severity.
 |
Table 5 Correlation Among CDI Severities, Genotypes and ermB of C. difficile with Resistance to MLSB |
Discussion
CDI is increasingly becoming a public health concern worldwide. During recent years, several studies conducted in China have revealed that CDI might gradually be a leading intestinal infection related to antibiotic usage. Furthermore, more attention should be paid to the antibiotic resistance of C. difficile in China, following the clarification of the molecular epidemiology of CDI. To the best of our knowledge, this is the first study to disclose the molecular characteristics of toxigenic C. difficile resistant to MLSB in community acquired CDI patients in Eastern China to address the different genotypes resulting in various CDI severities. In this study, we found that C. difficile isolates in this region have a high level of resistance to MLSB, with 88.4% resistance to CLI and 77.9% resistance to ERY. The A−B+ C. difficile isolates, especially ST37, led to more severe CDI, with higher severity score than that induced by A+B+ isolates in ermB-positive MLSB-resistant isolates, indicating that ermB-positive A−B+ C. difficile resistant to MLSB has become an epidemic clone inducing severe CDI in Eastern China.
The MLSB resistance rates in different countries ranged from 14.7% to 90.9% between 2002 and 2009.17 The strains of C. difficile carried over 50% of MLSB resistance rates with high-level resistance in Asia, and the MIC90 (mg/L) was equal to or more than 128 mg/L, except in a study in Taiwan.26 Antibiotic resistance data for C. difficile strains are still not enough to reveal the accurate antibiotic resistance profile in China. MLSB resistance data over the last 10 years showed that the average rate of resistance to ERY and CLI was 80.2% and 81.7% in China mainland, respectively. Our previous study results showed that the MLSB resistance rates in Zhejiang were lower than those observed in Shanghai.15,27 Nevertheless, high levels of MLSB with 88.4% (168/190) resistance to CLI and 77.9% (148/190) resistance to ERY were observed in this study, and 100% (88/88) of the A−B+ C. difficile isolates were resistant to MLSB. These data show a sharp increase in MLSB resistance with statistical differences in comparison with the data on ERY (64.7%, 266/411) and CLI (62.5%, 257/411) resistance obtained in our previous study before 2015 (ERY: χ2 = 10.52, P = 0.001, and CLI: χ2 = 42.06, P < 0.001).15 Thus, an obvious elevated trend of MLSB resistance was observed, which is noteworthy.
It has been well documented that high-level resistance against MLSB requires the ermB gene despite the low carrying rate of the ermB gene commonly found in MLSB-resistant C. difficile strains.17 A high proportion, that is, 85% of ermB-positive A−B+ C. difficile isolates presented resistance to MLSB antibiotics in a previous study.28 A study on the emergence of an epidemic due to a CLI-resistant C. difficile clone among Polish patients also showed that both A−B+ and A+B+ strains were resistant to MLSB antibiotics and that it was related with the ermB gene.13 Interestingly, we also found that a remarkably high proportion, that is, 90% (81 of 88) of A−B+ and 86.7% (78 of 90) of A+B+ C. difficile isolates showed resistance to MLSB associated with the presence of ermB. The above results demonstrated that C. difficile strains resistant to MLSB might be mainly driven through the ermB gene in Eastern China. We are currently conducting a large-scale molecular epidemiological study to determine whether the ermB gene mediates MLSB resistance.
PCR ribotypes 027 and 078 were hypervirulent C. difficile inducing severe CDI. We found that patients infected by the ermB-positive A−B+ isolates resistant to MLSB had more severe symptoms, with higher CDI severity scores than those of patients infected by A+B+ isolates. Interestingly, the ermB-positive ST37 strain, one of the three A−B+ genotypes in this study, seemed to be the main genotype driving moderate-to-severe CDI. Moreover, all the CDI patients were community-acquired with high diverse STs, demonstrating that community-associated CDI might be a main pattern of C. difficile transmission in China. The above similar conclusions were drawn from our previous studies.15,29 A high proportion of MLSB resistance among A−B+ isolates, especially ST37, should be underlined in patients with community-acquired CDI in comparison with that of A+B+ C. difficile. Resistance to MLSB enhanced the risk of CDI and promoted the persistence and transmission of C. difficile. The ermB gene is located in a transposon incorporated at a homologous site to the tcdA gene, indicating that there is a direct association between the biological characteristics of A−B+ isolates and MLSB resistance.28 However, it is still not clear why only ST37 rather than ST39 and ST81 caused moderate to severe CDI. We speculate that gene polymorphism, interaction among intestinal bacteria, metabolites, and other factors might have affected the pathogenicity of ST37.
Epidemics of CDI induced by MLSB-resistant C. difficile have been reported in the US and Europe.11,30,31 PCR ribotype 027 resistant to MLSB has been considered a significant and alarming emergence, driving the spread of this clone. The multiple locus variable number tandem repeat analysis showed that CLI-resistant ribotype 027 has a distant genetic relationship with CLI-susceptible 027 with a summed tandem repeat difference of 17.11 In the present study, MLST was used to analyze the genetic relationship among different isolates because of various STs existed. The results revealed that 93.7% of the isolates, including all the A−B+ and 88.2% of A+B+ isolates, were resistant to MLSB. Most C. difficile strains have obtained the characteristic of MLSB resistance with the overwhelming trend. The above results strongly revealed that C. difficile with MLSB resistance has been gradually becoming a dominant resistance phenotype in Eastern China. However, our study still has its own limitations as follows. This study enrolled a small scale of C. difficile isolates from only two tertiary hospitals, and only CA CDI cases were involved in this study. Moreover, we did not test resistance to other antibiotics, including fluoroquinolones, aminoglycoside, and tetracylines; therefore, the dynamic change of this clone should be underlined in the future in a large-scale study on molecular epidemiology of CDI.
Conclusions
In conclusion, the results of the present study showed the latest MLSB resistance development of C. difficile isolates from CA CDI patients in Eastern China. MLSB-resistant C. difficile isolates have been increasing in CDI cases dynamically. The ermB-positive A−B+ C. difficile resistant to MLSB has been a potential epidemic clone inducing severe CDI in this region. The epidemic scale of this clone might be widened gradually, and we speculate that MLSB resistance in A−B+ C. difficile with the ermB gene might be a probable predictor of enhanced CDI. Continuous surveillance by genotyping and antibiotic resistance testing focusing on ermB-positive A−B+ C. difficile resistant to MLSB should be conducted for monitoring changes in MLSB resistance and preventing epidemics with severe CDI.
Ethics Approval
The ethics committee of Zhejiang Provincial Center for Disease Control and Prevention has approved this study.
Abbreviations
MLSB: Macrolide-lincosamide-streptogramin B; Clostridioides difficile infection: CDI; ermB: erythromycin resistance methylase gene; CLI: Clindamycin; ERY: erythromycin; HFPH: Affiliated Hangzhou First People’s Hospital; LSPH: Lishui Second People’s Hospital; SHEA/IDSA: Society for Healthcare Epidemiology of America and the Infectious Diseases Society of America; ZJCDC: Zhejiang Provincial Center for Disease Control and Prevention; S-RBCs: Stool-red blood cells; S-WBCs: Stool-white blood cells; OB: Occult blood; CRP: C-reactive protein; WBCs: White-blood cells; Hb: hemoglobin; PLT: Platelets; MIC: Minimal inhibitory concentration; CLSI: Clinical and Laboratory Standards Institute; MLST: Multilocus sequence typing; ST: Sequence type.
Acknowledgment
We would like to thank Editage for English language editing.
Author Contributions
LYZ, YL, and DZJ wrote the manuscript and performed antibiotics resistance testing. QB, YL, YWT and JMJ analyzed the data. LYZ, LQW and XJW collected clinical samples and summarized clinical data. LYZ, YL, JLY and XJS performed isolation, MLST and antibiotics resistance testing. DZJ, YWT, and JMJ conceived the study and designed the experiments. XJW and YWT edited the manuscript. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work. All authors also read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81471998), the Key Research and Development Program of Zhejiang (2015C03048) and the Medical Health and Technology Plan of Zhejiang (2017KY289).
Disclosure
Yi-Wei Tang was an employee of Cephei, Danaher Diagnostic Platform, Shanghai, China during the conduct of the study. All authors declare that they have no other competing interests.
References
1. Bacci S, Molbak K, Kjeldsen MK, Olsen KE. Binary toxin and death after Clostridium difficile infection. Emerg Infect Dis. 2011;17(6):976–982. doi:10.3201/eid/1706.101483
2. Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7(7):526–536. doi:10.1038/nrmicro2164
3. Chang JY, Antonopoulos DA, Kalra A, et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile—associated diarrhea. J Infect Dis. 2008;197(3):435–438. doi:10.1086/586912
4. Jangi S, Lamont JT. Asymptomatic colonization by Clostridium difficile in infants: implications for disease in later life. J Pediatr Gastroenterol Nutr. 2010;51(1):2–7. doi:10.1097/MPG.0b013e3181d29767
5. Solomon K, Martin AJ, O’Donoghue C, et al. Mortality in patients with Clostridium difficile infection correlates with host pro-inflammatory and humoral immune responses. J Med Microbiol. 2013;62(Pt 9):1453–1460. doi:10.1099/jmm.0.058479-0
6. Brandt LJ. American journal of gastroenterology lecture: intestinal microbiota and the role of fecal microbiota transplant (FMT) in treatment of C. difficile infection. Am J Gastroenterol. 2013;108(2):177–185. doi:10.1038/ajg.2012.450
7. Simor AE. Diagnosis, management, and prevention of Clostridium difficile infection in long-term care facilities: a review. J Am Geriatr Soc. 2010;58(8):1556–1564. doi:10.1111/j.1532-5415.2010.02958.x
8. Vedantam G, Clark A, Chu M, McQuade R, Mallozzi M, Viswanathan VK. Clostridium difficile infection: toxins and non-toxin virulence factors, and their contributions to disease establishment and host response. Gut Microbes. 2012;3(2):121–134. doi:10.4161/gmic.19399
9. Cohen SH, Gerding DN, Johnson S, et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA). Infect Control Hosp Epidemiol. 2010;31(5):431–455. doi:10.1086/651706
10. Peng Z, Jin D. Update on antimicrobial resistance in Clostridium difficile: resistance mechanisms and antimicrobial susceptibility testing. J Clin Microbiol. 2017;55(7):1998–2008. doi:10.1128/JCM.02250-16
11. Drudy D, Goorhuis B, Bakker D, et al. Clindamycin-resistant clone of Clostridium difficile PCR ribotype 027, Europe. Emerg Infect Dis. 2008;14(9):1485–1487. doi:10.3201/eid1409.071346
12. Kuijper EJ, Barbut F, Brazier JS, et al. Update of Clostridium difficile infection due to PCR ribotype 027 in Europe, 2008. Euro Surveillance. 2008;13(31):18942.
13. Pituch H, Van Belkum A, Van Den Braak N, et al. Recent emergence of an epidemic clindamycin-resistant clone of Clostridium difficile among Polish patients with C. difficile-associated diarrhea. J Clin Microbiol. 2003;41(9):4184–4187. doi:10.1128/JCM.41.9.4184-4187.2003
14. Tang C, Cui L, Xu Y, et al. The incidence and drug resistance of Clostridium difficile infection in Mainland China: a systematic review and meta-analysis. Sci Rep. 2016;6:37865. doi:10.1038/srep37865
15. Jin D, Luo Y, Huang C, et al. Molecular epidemiology of clostridium difficile infection in hospitalized patients in Eastern China. J Clin Microbiol. 2017;55(3):801–810. doi:10.1128/JCM.01898-16
16. Ackermann G, Degner A, Cohen SH, Silva J
17. Huang H, Weintraub A, Fang H, Nord CE. Antimicrobial resistance in. Int J Antimicrob Agents. 2009;34(6):516–522. doi:10.1016/j.ijantimicag.2009.09.012
18. (CLSI) CaLSI. Defining, establishing, and verifying reference intervals in the clinical laboratory: approved Guideline-Third Edition. 2010.
19. Ichihara KBJ. IFCC committee on reference intervals and decision limits (C-RIDL). An appraisal of statistical proceduces used in derivation of reference intervals. Clin Chem Lab Med. 2010;48:1537–1551. doi:10.1515/CCLM.2010.319
20. Huang B, Jin D, Zhang J, et al. Real-time cellular analysis coupled with a specimen enrichment accurately detects and quantifies Clostridium difficile toxins in stool. J Clin Microbiol. 2014;52(4):1105–1111. doi:10.1128/JCM.02601-13
21. Ryder AB, Huang Y, Li H, et al. Assessment of Clostridium difficile infections by quantitative detection of tcdB toxin by use of a real-time cell analysis system. J Clin Microbiol. 2010;48(11):4129–4134. doi:10.1128/JCM.01104-10
22. McDonald LC, Killgore GE, Thompson A, et al. An epidemic, toxin gene–variant strain of Clostridium difficile. N Eng J Med. 2005;353(23):2433–2441. doi:10.1056/NEJMoa051590
23. Griffiths D, Fawley W, Kachrimanidou M, et al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol. 2010;48(3):770. doi:10.1128/JCM.01796-09
24. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. National Committee for Clinical and Laboratory Standards Wayne; 2017.
25. He M, Miyajima F, Roberts P, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 2013;45(1):109–113. doi:10.1038/ng.2478
26. Collins DA, Hawkey PM, Riley TV. Epidemiology of Clostridium difficile infection in Asia. Antimicrob Resist Infect Control. 2013;2(1):21. doi:10.1186/2047-2994-2-21
27. Dong D, Peng Y, Zhang L, Jiang C, Wang X, Mao E. Clinical and microbiological characterization of Clostridium difficile infection in a tertiary care hospital in Shanghai, China. Chin Med J (Engl). 2014;127(9):1601–1607.
28. Van den Berg RJ, Claas EC, Oyib DH, et al. Characterization of toxin A-negative, toxin B-positive Clostridium difficile isolates from outbreaks in different countries by amplified fragment length polymorphism and PCR ribotyping. J Clin Microbiol. 2004;42(3):1035–1041. doi:10.1128/JCM.42.3.1035-1041.2004
29. Zheng Y, Luo Y, Lv Y, et al. Clostridium difficile colonization in preoperative colorectal cancer patients. Oncotarget. 2017;8(7):11877–11886. doi:10.18632/oncotarget.14424
30. Johnson S, Samore MH, Farrow KA, et al. Epidemics of diarrhea caused by a clindamycin-resistant strain of Clostridium difficile in four hospitals. N Eng J Med. 1999;341(22):1645–1651. doi:10.1056/NEJM199911253412203
31. Kuijper EJ, de Weerdt J, Kato H, et al. Nosocomial outbreak of Clostridium difficile-associated diarrhoea due to a clindamycin-resistant enterotoxin A-negative strain. Euro J Clin Microbiol & Infect Dis. 2001;20(8):528–534. doi:10.1007/s100960100550
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
