Back to Journals » International Journal of Nanomedicine » Volume 9 » Issue 1
High gene delivery efficiency of alkylated low-molecular-weight polyethylenimine through gemini surfactant-like effect
Authors Liu S , Huang W, Jin M, Wang Q, Zhang G, Wang X , Shao S, Gao Z
Received 21 March 2014
Accepted for publication 27 April 2014
Published 28 July 2014 Volume 2014:9(1) Pages 3567—3581
DOI https://doi.org/10.2147/IJN.S64554
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Shan Liu,1,2 Wei Huang,1,2 Ming-Ji Jin,1,2 Qi-Ming Wang,1,2 Gan-Lin Zhang,3 Xiao-Min Wang,3 Shuai Shao,1,2 Zhong-Gao Gao1,2
1State Key Laboratory of Bioactive Substance and Function of Natural Medicines, 2Department of Pharmaceutics, Institute of Materia Medica, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, People’s Republic of China; 3Beijing Hospital of Traditional Chinese Medicine affiliated to Capital Medical University, Beijing, People’s Republic of China
Abstract: To our knowledge, the mechanism underlying the high transfection efficiency of alkylated low-molecular-weight polyethylenimine (PEI) is not yet well understood. In this work, we grafted branched PEI (molecular weight of 1,800 Da; bPEI1800) with lauryl chains (C12), and found that bPEI1800-C12 was structurally similar to gemini surfactant and could similarly assemble into micelle-like particles. Stability, cellular uptake, and lysosome escape ability of bPEI1800-C12/DNA polyplexes were all greatly enhanced after C12 grafting. bPEI1800-C12/DNA polyplexes exhibited significantly higher transfection efficiency than Lipofectamine™ 2000 in the presence of serum. Bioluminescence imaging showed that systemic injection of bPEI1800-C12/DNA polyplexes resulted in intensive luciferase expression in vivo and bioluminescence signals that could be detected even in the head. Altogether, the high transfection efficacy of bPEI1800-C12 was because bPEI1800-C12, being an analog of gemini surfactant, facilitated lysosome escape and induced the coil–globule transition of DNA to assemble into a highly organized micelle-like structure that showed high stability.
Keywords: self-organization, alkylation, luciferase, bioluminescence imaging
Introduction
Gene therapy is essentially based on the development of efficient and safe gene carrier systems able to transfer plasmid DNA or small interfering RNA (siRNA) into cells.1 Among the various nonviral systems, polyethyleneimine (PEI) is one of the most intensively studied systems due to its high transfection efficiency.2,3 Unfortunately, the high transfer efficiency of PEI is usually accompanied by high toxicity, so various means of structure modification have been tried in order to obtain a good compromise between efficiency and toxicity for PEI, such as poly(lactic-co-glycolic acid)-linear PEI with a molecular weight of 22,000 Da (lPEI22K);4 branched PEI (bPEI)-g-ovalbumin with a molecular weight of 600 Da (bPEI0.6K);5 heparin-grafted bPEI with a molecular weight of 1,800 Da (bPEI1.8K);6 hyaluronic acid-grafted bPEI with a molecular weight of 25,000 Da (bPEI25K);7 polyethylene glycol-g-bPEI with a molecular weight of 25,000 Da (bPEI25K);8 and hydrophobic grafting (cholesteryl, cholanic acid, fatty acid residues, and hydrophobic chains).9 Among the many modification methods of PEI, grafting PEI with hydrophobic moieties has proved to be an effective way of obtaining a favorable tradeoff between efficacy and toxicity, by: 1) increasing the transfection efficiency by elevated lipophilicity, which enhances the affinity of PEI to biomembranes and facilitates the cellular entry;10 2) improving the stability of polyplexes against serum and DNAase;11 and 3) lowering the toxicity by reduced charge density, which causes less cell damage.12
It has been demonstrated that hydrophobically modified low-molecular-weight (LMW) PEI is not only an efficient gene vector itself,13,14 but can also act as an intermediate in the fabrication of hybrid multifunctional carriers for gene delivery.15–19 To our knowledge, however, the mechanism underlying the high transfection efficacy of hydrophobically modified LMW PEI is not yet well understood. The high transfection efficiency of PEI is widely considered to be due to its high charge density and the proton sponge effect, which triggers the escape of PEI/DNA polyplexes from lysosomes. It is believed that LMW PEIs, due to their low branching degree, cannot compact DNA efficiently, and such low compaction ability would become even lower were they conjugated with hydrophobic moieties. Likewise, it has been proved that LMW PEI itself could not evoke efficient endosomal escape by the proton sponge effect.20 Thus, in the case of hydrophobically modified LMW PEI, it seems that neither positive charge density potential nor the proton sponge effect contributes to its high transfection efficiency.
It is widely assumed that escape of PEI/DNA polyplexes from lysosomes is triggered by the proton sponge effect. So far, however, the proton sponge hypothesis has not been proved by experiments, and this hypothesis has been recently challenged.21 Besides the proton sponge effect, other effects, such as surfactant activity,22 are believed to contribute to the lysosomal membrane permeabilization capacity of PEI23 and cause lysosome release of polyplexes. It was found that cationic surfactant could complex with DNA, resulting in a micelle-like structure that precluded the aggregation of polyplexes, and such cationic surfactant could also destabilize the endosomal membrane and trigger DNA release into the cytoplasm.24 Gemini surfactant, a recently extensively investigated gene vector,25 was also found to be efficient at facilitating biomembrane penetration and endosomal escape.26 Hydrophobic grafting of bPEI would give rise to cationic amphiphile bPEI derivatives, which are structurally similar to cationic surfactant or gemini surfactant, all of which possess cationic head groups and hydrophobic tails. Moreover, it was found that micelle-forming structures played a key role in enhancing transfection of lipid-modified bPEI (molecular weight: 1,800 Da [bPEI1800]),27 so we speculated that the structure of hydrophobically modified LMW PEI might be highly related to its transfection efficiency.
In light of the aforementioned information, we aimed to investigate the structure/activity relationship of alkylated LMW PEI. Hydrocarbon chains (C12) were used to graft LMW PEI (bPEI1800) and a series of cationic amphiphile PEI derivatives (bPEI1800-C12) were obtained. The structure, physicochemical characteristics, interactions with cells, in vitro transfection, and, especially, the in vivo gene delivery efficacy of bPEI1800-C12 were fully investigated. The obtained results suggested that bPEI1800-C12 could assemble into highly stable micelle-like particles and that they triggered the coil–globule transition of DNA similarly to cationic surfactant and possessed high gene delivery efficacy in vitro and in vivo.
Materials and methods
Materials and cells
All reagents and solvents were used as received without further purification. bPEI1800 was purchased from Alfa Aesar (Ward Hill, MA, USA). 1-bromododecane, heparin sodium salt (150 USP [United States Pharmacopeia] units/mg), N-(2-hydroxyethyl) piperazine-N’-ethanesulfonic acid (HEPES), fluorescein isothiocyanate (FITC) isomer, ethidium bromide, chlorpromazine, methyl-β-cyclodextrin (M-β-CD), and sodium azide were obtained from Sigma-Aldrich Co. (St Louis, MO, USA). Trypsin, fetal bovine serum (FBS), Dulbecco’s Modified Eagle’s Medium (DMEM), and Roswell Park Memorial Institute (RPMI) 1640 medium were all provided by GE Healthcare UK Ltd (Little Chalfont, UK). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was from AMRESCO LLC (Solon, OH, USA). LysoTracker® Red dye, Hoechst 33258, apoptosis detection kit (C1062), and Bradford protein assay kit (P0006) were all from Beyotime Institute of Biotechnology (Shanghai, People’s Republic of China). Luciferase assay kit (E1500) was from Promega Corporation (Fitchburg, WI, USA). Other chemicals and reagents were of analytical grade. Mouse mammary tumor cell line 4T1 was provided by the Department of Pathology in Institute of Medicinal Biotechnology in Peking Union Medical College (Beijing, People’s Republic of China) and cultured in RPMI 1640 containing 10% FBS.
Synthesis of bPEI1800-C12 derivatives
Grafting bPEI1800 with hydrophobic C12 chains was carried out as reported previously.28 Briefly, 1-bromododecane was reacted with a solution of bPEI1800 (1 g, 0.56 mmol) in dichloromethane (150 mL) at room temperature (RT) for 24 hours. The resulting solvent was concentrated under reduced pressure and further dialyzed, first against an ethanol/water (1:1) solution (4 L five times), then against double-distilled water for 1 day. The mixture was concentrated and lyophilized. Proton nuclear magnetic resonance (1H NMR) spectra of bPEI1800 and its derivatives were recorded on a 400 MHz Mercury plus instrument (Varian Inc., Palo Alto, CA, USA), and substitution degree was also calculated based on the integration of 1H NMR. FITC fluorescent labeling of the polymers was performed as reported previously.29
pH titration
pH titrations were performed as reported previously.30 In short, 2 mL of each PEI solution (113 mM in −CH2CH2NH− units) were adjusted to pH 12 with NaOH. Five μL amounts of 1 M HCl were added sequentially, and the pH after each addition was measured; a solution of NaCl (113 mM) was titrated similarly as a control.
Gel retardation assay
Agarose gel retardation assays were conducted following standard procedure. Varying concentrations of PEI or PEI derivatives and equal volume (10 μL) of plasmid DNA solutions (concentration kept at 20 ng/μL) were mixed in HEPES-buffered saline ([HBS] 10 mM HEPES, 150 mM NaCl, pH 7.4), respectively, incubated at RT for 30 minutes, then mixed with 4 μL of 6× loading buffer, loaded into 1.0% agarose gel (in Tris-acetate-ethylenediaminetetraacetic acid [TAE] buffer), and run for 60 minutes at 80 V. After staining of the agarose gel with 0.5 μg/mL ethidium bromide for 30 minutes, the plasmid DNA bands were visualized at 254 nm with an ultraviolet gel imaging system (BIO- BEST 135A; SIM International Group Co., Ltd, Los Angeles, CA, USA).
Morphology
The morphology of bPEI/DNA polyplexes was visualized by transmission electron microscopy (TEM). Briefly, 20 μL of 20 ng/μL DNA was mixed and incubated with an equal volume of bPEI solution in HBS (pH 7.4) for 30 minutes at RT and diluted with 80 μL HBS (pH 7.4). Ten microliters of the diluted polyplexes solution was dropped onto a copper grid coated with carbon membrane, then the copper grid was air-dried and negatively stained with 1% sodium phosphotungstate. Subsequently, the morphology of polyplexes was observed using a transmission electron microscope (Hitachi H-7650; Hitachi Ltd., Tokyo, Japan) with an acceleration voltage of 80 kV.
Heparin displacement assay
Ten microliters of polyplex solution in HBS (pH 7.4), prepared according to the method described in the Gel retardation assay section, were exposed to 5 μL of HBS (pH 7.4) containing serial concentrations of heparin sodium salt for 1 hour at RT. Then, the mixture was run in 1% agarose gel as described.
Hemolysis assay
Rat erythrocytes were collected from heparin-treated blood and washed with 0.01 M isotonic phosphate-buffered saline ([PBS] pH 7.4) four times (700 g for 10 minutes at 4°C). The erythrocyte pellet was diluted tenfold with 0.01 M isotonic PBS (pH 7.4) to a concentration of 109/mL. A 75 μL aliquot was added to a 96-well plate containing 75 μL serial solutions of bPEI1800 or its derivatives in the corresponding buffer. The final concentrations of bPEI1800 and its derivatives varied from 6 μg/mL to 100 μg/mL. After incubation for 1 hour at 37°C with constant shaking, unlysed erythrocytes were removed by centrifugation (700 g for 10 minutes at 4°C), then 80 μL supernatant was transferred to a new 96-well plate and hemoglobin absorption was measured at 450 nm using a SpectraMax 190 Absorbance Microplate Reader (Molecular Devices LLC, Sunnyvale, CA, USA). Triton™ X-100 (1%) (Sigma-Aldrich Co.) was used as the positive control (100% lysis). Experiments were performed in triplicate.
Zeta potential measurement
Polyplexes were prepared at ratios from 1:1 to 20:1 (polymer to DNA, weight [w]/w) using the method described in the Gel retardation assay section, then diluted with HBS (pH 7.4), and the final DNA concentration was 0.5 μg/mL. The zeta potential of each sample was measured by a Nicomp 380 ZLS analyzer (Particle Sizing Systems, Port Richey, FL, USA). Experiments were performed in triplicate.
Cytotoxicity
Cytotoxicity of bPEI1800-C12 was evaluated by MTT assay as previously reported.31 Briefly, 4T1 cells were seeded onto 96-well plates at a density of 4×103/well. After incubation overnight for adherence, the previous medium was replaced with FBS-supplemented medium containing various concentrations of bPEI1800 or bPEI1800-C12. After 20 or 44 hours of incubation, the medium was removed and 100 μL fresh medium containing 0.5 mg/mL MTT was added to each well. Cells were further incubated for 4 hours, and medium was replaced with 150 μL dimethyl sulfoxide to solubilize the converted formazan. Absorbance was measured at 570 nm with the SpectraMax 190 Absorbance Microplate Reader. Cells without exposure to the polymers were used to represent 100% cell viability. Experiments were performed in triplicate.
Cellular uptake studies
For confocal laser scanning microscope visualization, 4T1 cells were seeded onto a 24-well plate with sterile glass slides at a density of 4×104 cells/well and incubated overnight for attachment. Then 4T1 cells were rinsed thrice with serum-free medium and incubated with polyplexes prepared with FITC-labeled polymers for 4 or 24 hours at 37°C: the first 4-hour incubation was in serum-free medium; for longer incubation, medium with serum was supplemented. At the determined time point, cells were washed thrice with PBS and the cell nuclei were stained with Hoechst 33258 before observation with a confocal scanning laser microscope (TCS SP2; Leica Microsystems, Wetzlar, Germany).
Endocytosis pathway
For endocytosis pathway studies, 4T1 cells were seeded onto 12-well plates until grown to 85% confluence, rinsed thrice with serum-free medium, and preincubated in serum-free medium containing 10 μg/mL chlorpromazine, 5 mM M-β-CD, or 10 mM sodium azide for 1 hour. Then, the previous medium was removed and supplemented with fresh serum-free medium that contained chemical pathway inhibitors (10 μg/mL chlorpromazine, 5 mM M-β-CD, or 10 mM sodium azide) and polyplexes prepared with FITC-labeled polymers. Incubation was continued for 4 hours. Then, cells were washed thrice with PBS, detached by trypsin, centrifuged for 5 minutes, resuspended, and analyzed with a FACSCaliber™ flow cytometer (BD, Franklin Lakes, NJ, USA). The intracellular amount of polyplexes was represented by the relative fluorescence intensity, and the group without treatment, in which mean fluorescence intensity was determined to be 100%, was set as the control.32
Intracellular trafficking
Intracellular trafficking studies were performed similarly to cellular uptake studies. 4T1 cells were similarly seeded, attached, treated with FITC-labeled polyplexes, and visualized, except that, after PBS washing, cells were further treated with LysoTracker Red and Hoechst according to the manufacturer’s protocol.
In vitro transfection
In vitro transfection studies were performed with plasmid GL 4.50 (pGL 4.50) and plasmid EGFP (pEGFP), which encoded luciferase and green fluorescence protein, respectively. 4T1 cells were seeded onto a 96-well plate at a density of 4×103 cells/well and grown for 24 hours. Then, medium was replaced with 40 μL serum-containing medium, and 10 μL of freshly prepared polyplexes in HBS containing 100 ng DNA was added to each well. lPEI22K and Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA) were used as the controls. The transfection of lPEI22K was carried out similarly to that of the bPEI1800, except that the weight ratio was set at 5:1 (polymer to DNA), and the transfection experiments with Lipofectamine 2000 were performed according to the manufacturer’s instructions. After 4 hours of transfection, 125 μL of 10% FBS medium was added to each well, and the cells were incubated for an additional 20 hours. Finally, cells were washed twice with PBS, luciferase activity was detected according to standard protocol for the luciferase assay system, and the protein concentration of each well was measured using the Bradford protein assay kit.
In vivo gene delivery
Animal experiments (BALB/c mouse, female, 6-8 weeks old) adhered to the Principles of Laboratory Animal Ethics Committee of the Institute of Materia Medica in Peking Union Medical College, and were conducted with necessary humane care. Five hundred microliters of 1,000 ng/μL bPEI1800 and bPEI1800-C12-19.72 (bPEI1800-C12 where the substitution degree of C12 was 19.72) in HBS were separately mixed with an equal volume of 200 ng/μL pGL 4.50 solutions, and the resulting polyplexes were incubated at RT for 30 minutes before being injected into mice through the tail vein (200 μL polyplexes solution containing 20 μg plasmid DNA for each mouse). Naked plasmid DNA solution was used as the control. Twenty-four hours post-injection, mice were anesthetized with isoflurane, and imaging began 8 minutes after administration of D-luciferin. Bioluminescence imaging was performed with an IVIS® Spectrum in vivo imaging system (IVISSPE; PerkinElmer, Inc., Waltham, MA, U.S.) and the images were analyzed by Living Image software (version 4.3.1; PerkinElmer, Inc.). Anesthesia was maintained during imaging and images were acquired for 2 minutes. After bioluminescence imaging, the mice were quickly killed by cervical dislocation; their livers, hearts, spleens, lungs, kidneys, and brains were collected, washed with normal saline, and suspended in 800 μL (liver, brain) or 400 μL (heart, spleen, lung, kidney) lysis buffer consisting of passive lysis buffer, phenylmethylsulfonyl fluoride, and protease inhibitor mixture,30 homogenized by probe sonication for 30 seconds, and centrifuged (4°C, 20,000× g, 10 minutes). Luciferase activity of the supernatants was detected according to the standard protocol for the luciferase assay system and the protein concentration was measured by the Bradford protein assay kit.
Statistical analysis
Results were expressed as mean ± standard deviation of at least three independent experiments. Student’s t-test was used to evaluate the difference between two groups. One-way analysis of variance (ANOVA) with Bonferroni’s post hoc test was used to compare the statistical significance among the groups, and P<0.05 was considered significant.
Results
Synthesis of bPEI1800-C12 derivatives
1-bromododecane was chosen as C12 chain donor for the hydrophobic grafting of bPEI1800. Characteristic chemical shift (δ) of 0.92 (−CH3) and 1.32 (−(CH2)11−) in the 1H NMR spectrum of bPEI1800-C12 indicated the successful grafting of C12 (Figure S1). By varying the weight ratios of 1-bromododecane and bPEI1800, we obtained bPEI1800 derivatives of different substitution degrees which were calculated by integration of 1H NMR. Since the molecular formula of bPEI1800-C12 is (C2H4N)42(C12H25)42n, the degree of C12 substitution was calculated by the following equation:
25n/4=(peak area of −C12H25)/(peak area of −C2H4N−) | (1) |
where n is the substitution degree of C12. The calculated substitution degrees (6.08, 13.44, 17.12, and 22.72) were quite close to their theoretical values (5.28, 12.5, 19.72, and 25), so the bPEI1800 derivatives were abbreviated to bPEI1800-C12-5.28, bPEI1800-C12-12.5, bPEI1800-C12-19.72, and bPEI1800-C12-25, respectively.
Buffering capacity of bPEI1800-C12
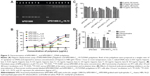
Since the high buffering capacity of PEI is widely considered as a guarantee for efficient endosomal escape of polyplexes and high transfection efficiency, we investigated the influence of C12 grafting on the buffering capacity of bPEI via pH titration. Buffering capacity of bPEI1800-C12 derivatives was markedly reduced compared with that of bPEI1800 (Figure 1A). After C12 grafting, partial primary amines of PEI turned into secondary amines and thus led to reduced protonation ability. Additionally, grafting bPEI1800 with C12 caused strong hydrophobic repulsion in bPEI1800-C12 molecules, and such hydrophobic repulsion rearranged amine groups of bPEI1800-C12, resulting in decreased amine spacing, which led to retarded protonation and reduced buffering capability.33
DNA condensation of bPEI1800-C12 via retardation assay
As shown in Figure 1B, bPEI1800 could completely condense DNA at a weight ratio of 0.4:1 (polymer to DNA), while, in the case of bPEI1800-C12 derivatives, with the increase of C12 substitution degree, weight ratios for complete DNA condensation were 0.6:1, 0.8:1, or 1:1 (polymer to DNA, w/w), suggesting that C12 grafting reduced the binding ability of bPEI1800-C12 to DNA, and the binding ability of bPEI1800-C12 decreased with the increment of substitution degree. The reasons that bPEI1800-C12 exhibited less efficient binding to DNA compared with the parent bPEI1800 is that electrostatic interaction between PEI and DNA is the driving force for compaction and that C12 grafting retarded the protonation of bPEI1800-C12 (Figure 1A).
Morphology of bPEI1800-C12/DNA polyplexes
bPEI1800-C12 polymers at a concentration of 4 mg/mL showed the appearance of light blue opalescence (Figure S2), indicating that bPEI1800-C12 self-organized into nano-sized particles in water solution. TEM visualization showed that bPEI1800-C12-19.72/DNA polyplexes exhibited a micelle-like structure at pH 7.4 (Figure 2B), which consisted of a hard inner core and outer shell (about 300 nm in diameter). bPEI1800/DNA polyplexes displayed a plain solid spherical structure ([Figure 2A] about 180 nm in diameter) at pH 7.4. With the synthesis route of the present study, C12 chains were mostly grafted to the primary amino groups of bPEI1800, and such primarily grafted C12 chains were believed to locate around the periphery of bPEI1800.13 In order to gain thermodynamic stability, bPEI1800-C12 should rationally self-assemble into a micelle-like structure in aqueous media, with C12 chains forming the hydrophobic inner core and bPEI moiety composing the outer shell (Figure 2C). When complexing DNA with bPEI1800-C12 at pH 7.4, DNA underwent coil–globule transition34 and was bound to the exterior bPEI moiety of the bPEI1800-C12 micelle (Figure 2C), forming multi-hydrophobic domains24 along the DNA chain.
In vitro stability of bPEI1800-C12/DNA polyplexes
Heparin displacement assay was conducted to test the stability of polyplexes. bPEI1800/DNA and bPEI1800-C12-19.72/DNA polyplexes prepared at a weight ratio of 3:1 (polymer to DNA) were separately exposed to heparin in HBS (pH 7.4) for 1 hour at RT. Results from the subsequent gel electrophoresis (Figure 3A) indicated that DNA in bPEI1800/DNA polyplexes was completely displaced by heparin at a concentration of 0.25 mg/mL, but bPEI1800-C12-19.72/DNA polyplexes were quite robust even when the concentration of heparin was up to 4 mg/mL, suggesting that C12 grafting offered bPEI1800-C12/DNA polyplexes high stability against anion displacement. Since there are many polyanions in serum that are apt to cause instability of polyplexes, the robustness of bPEI1800-C12/DNA polyplexes against anion displacement suggested its potential for in vivo application. Additionally, zeta potentials of all polyplexes prepared with bPEI1800-C12 were much higher than those of the polyplexes prepared with bPEI1800 (Figure S3) at the same weight ratio (polymer to DNA), suggesting that bPEI1800-C12/DNA complexes maintained much higher stability in aqueous solution than bPEI1800/DNA complexes due to their highly organized structure.
Membrane destabilization activity of bPEI1800-C12
It has been reported that hydrocarbon chain-grafted PEI showed much stronger interaction with phospholipids than unmodified PEI,13 so we performed a hemolysis assay to study the interaction between biomembranes and bPEI1800-C12. bPEI1800-C12 polymers were incubated with erythrocytes at pH 7.4 at 37°C, and the released amount of hemoglobin was measured to represent membrane destabilization activity. As depicted in Figure 3B, bPEI1800 showed no membrane destabilization activity at concentrations up to 100 μg/mL,35 while bPEI1800-C12 imposed strong destabilization activity, and such destabilization activity increased with the increasing of both C12 substitution and concentration of bPEI1800-C12. The results obtained here indicate that bPEI1800-C12 showed great affinity to biomembranes and possessed fairly good membrane destabilization activity, which could help penetrate biomembranes and facilitate lysosome escape.
Cytotoxicity of bPEI1800-C12 on 4T1 cells
The cytotoxicity of bPEI1800-C12 on 4T1 cells was studied by MTT assay. As illustrated in Figure 3C, bPEI1800 showed no cytotoxicity on 4T1 cells after 24 and 48 hours’ incubation; 24-hour incubation of cells with 5 or 10 μg/mL bPEI1800-C12 polymers exerted no cytotoxicity; 5 or 10 μg/mL bPEI1800-C12 polymers were moderately toxic to 4T1 cells after 48 hours of treatment. The cytotoxicity of bPEI1800-C12 was probably due to membrane destabilization activity23 caused by membrane insertion of the free bPEI1800-C12, as demonstrated by hemolysis assay (Figure 3B).
Cellular uptake and endocytosis of bPEI1800-C12/DNA polyplexes
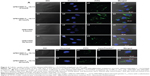
In order to investigate the cellular uptake of bPEI1800-C12/DNA polyplexes, FITC-labeled bPEI1800 and bPEI1800-C12 were complexed with DNA at a weight ratio of 3:1 (polymer to DNA), and their cellular uptake was visualized with confocal laser scanning microscopy. As depicted in Figure 4A, cellular uptake of both bPEI1800/DNA and bPEI1800-C12/DNA polyplexes was time-dependent, and cellular uptake of bPEI1800-C12/DNA polyplexes was dramatically higher than that of bPEI1800/DNA at the same time point.
Since bPEI1800-C12/DNA and bPEI1800/DNA polyplexes showed discriminated cellular uptake, we further studied their endocytosis using three kinds of chemical inhibitors of endocytosis:36 1) chlorpromazine, an inhibitor of clathrin-mediated endocytosis pathway; 2) M-β-CD, a cholesterol depletor for the inhibition of caveolae-mediated endocytosis; and 3) NaN3, which is used to inhibit the energy-dependent endocytosis pathway. The final concentrations of chlorpromazine, M-β-CD, and NaN3 used here were 10 μg/mL, 5 mM, and 10 mM, respectively, none of which showed cytotoxicity to 4T1 cells (Figure S4). As shown in Figure 3D, NaN3 treatment reduced the cellular uptake of bPEI1800/DNA polyplexes and bPEI1800-C12-19.72/DNA polyplexes by 37.8% and 61%, respectively, implying that both polyplexes entered cells by the combined processes of endocytosis and non-endocytosis. Only a small portion of bPEI1800/DNA polyplexes entered cells by energy-dependent pathways (37.8%), but most of the bPEI1800-C12-19.72/DNA polyplexes (61%) were internalized into cells through energy-dependent endocytosis. When treated with M-β-CD, cellular uptake of bPEI1800-C12-19.72/DNA polyplexes dropped by nearly 59.2%, which was close to the cellular uptake reduction rate of those treated with NaN3 (61%), suggesting that bPEI1800-C12-19.72/DNA polyplexes entered cells predominantly by caveolae-mediated endocytosis. Additionally, a previous report showed that particles larger than 200 nm in size were more likely to be internalized into cells through caveolae-mediated pathway.37 Hence, bPEI1800-C12/DNA polyplexes with bigger size and highly organized structure were more favorable to caveolae-mediated endocytosis. Additionally, the clathrin-mediated endocytic pathway also contributed to the endocytosis of bPEI1800-C12-19.72/DNA polyplexes, as proved by the 31.3% drop in cellular uptake caused by chlorpromazine.
Along with cellular uptake, intracellular trafficking is also an important issue for efficient delivery of DNA. Figure 4B illustrates the intracellular trafficking of bPEI1800-C12-19.72/DNA polyplexes after 4 and 24 hours’ incubation. We can see that only a few FITC-labeled bPEI1800-C12-19.72/DNA polyplexes (green fluorescence) were co-located in the lysosomes (red fluorescence), generating a yellow appearance due to the merge of green and red color (Figure 4Ba5 and b5), while most of the polyplexes were not situated in lysosomes but distributed throughout the cytoplasm, giving only green fluorescence (Figure 4Ba5 and b5). We have proved that most of the bPEI1800-C12-19.72/DNA polyplexes entered cells through caveolae-mediated endocytosis, which has been reported to be able to bypass lysosomes,36 so most of the bPEI1800-C12-19.72/DNA polyplexes could move directly to the nucleus without significant cellular obstacles.38 Moreover, although partial polyplexes could be trapped in the endosomes, some of them would have been released into the cytoplasm due to the membrane-destabilizing activity of the free bPEI1800-C12. As a result, only a few polyplexes that are not released into cytoplasm in time will end up being trapped in the lysosomes. Based on the above results, we concluded that C12 grafting not only greatly improved the cellular uptake of bPEI1800-C12-19.72/DNA polyplexes, but also altered their endocytosis pathways, leading to cellular trafficking without the obstacle of lysosomes, which was beneficial for the transfection of DNA.
In vitro transfection
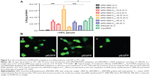
We also studied the transfection efficiency of bPEI1800-C12/DNA polyplexes on 4T1 cells using a luciferase reporter gene (pGL 4.50). The results depicted in Figure 5A indicate that all bPEI1800-C12/DNA polyplexes prepared at a weight ratio of 3:1 (polymer to DNA) showed higher luciferase activity than those prepared at a weight ratio of 5:1. Importantly, bPEI1800-C12-19.72/DNA at a weight ratio of 3:1 possessed the highest luciferase activity in serum-containing medium, which was 160-fold higher than that of bPEI1800 and significantly higher than that of lPEI22K (P<0.001) and Lipofectamine 2000 (P<0.05) under the same condition. Moreover, transfection efficacy of bPEI1800-C12-19.72 was also assessed using a green fluorescence protein reporter gene (pEGFP) on 4T1 cells in the presence of serum. Under the above optimized transfection condition (w/w =3, polymer to DNA), 4T1 cells exhibited strong green fluorescence 48 hours post-transfection with bPEI1800-C12-19.72/pEGFP polyplexes (Figure 5Ba), which was comparable to the green fluorescence of cells transfected with Lipofectamine 2000 (Figure 5Bb) and dramatically higher than that of cells transfected with bPEI1800 (Figure 5Bc). Such results were in line with the results of using pGL 4.50 as a reporter gene, suggesting that bPEI1800-C12-19.72 could deliver a variety of plasmid DNA types into cells and guarantee their successful expression as well.
In vivo transfection
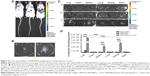
The fact that bPEI1800-C12-19.72/DNA polyplexes showed high stability against heparin and maintained higher in vitro transfection efficiency than Lipofectamine 2000 in the presence of serum indicated their great promise for in vivo application, so we studied the in vivo gene delivery efficiency of bPEI1800-C12-19.72 by bioluminescence imaging, and the in vivo distribution of bPEI1800-C12-19.72/DNA polyplexes was also investigated via luciferase activity measurement. Naked DNA, because of their low stability, cannot circulate into the systemic circulation and can only express locally at the injection site of the tail (Figure 6Aa). Compared to bPEI1800/DNA polyplexes (Figure 6Ab), bPEI1800-C12-19.72/DNA polyplexes showed dramatically stronger luminescence intensity in vivo (Figure 6Ac). Amazingly, luminescence signal could even be detected in the head after injection of bPEI1800-C12-19.72/DNA polyplexes (Figure 6Ac). Unfortunately, when the freshly dissected scalps and skulls of mice 24 hours post-injection with bPEI1800-C12-19.72/DNA polyplexes were imaged simultaneously, we found that luminescence signal in the head was actually located in the scalp rather than the skull (Figure 6B); this phenomenon was also confirmed by the result that neither luminescence signal (Figure 6Cc) nor luciferase expression (Figure 6D) were detected in the resected brain.
In vivo distribution studies revealed that naked DNA showed no luciferase expression in any of the detected organs (Figure 6Ca) and bPEI1800/DNA polyplexes expressed only in liver (Figure 6Cb), whereas bPEI1800-C12-19.72/DNA polyplexes possessed significantly higher expression in liver (P<0.001), spleen (P<0.001), and kidneys (P<0.001) (Figure 6Cc). The corresponding luciferase activity of each organ, shown in Figure 6D, was in accordance with the results of bioluminescence imaging (Figure 6C). As illustrated in Figure 6D, 24 hours after injection of bPEI1800-C12-19.72/DNA polyplexes, luciferase activity in liver was significantly higher than in spleen (P<0.001) and kidneys (P<0.001), and luciferase activity of all three organs (liver, spleen, kidneys) was significantly higher than in the corresponding organs of mice injected with bPEI1800/DNA polyplexes and naked DNA (P<0.001).
Discussion
As previously reported, grafting bPEI25K with acyl chains reduced the buffering capacity of PEI;10 herein, results from both pH titration and gel retardation assay led to the same conclusion, that C12 grafting lowered the compaction efficiency of bPEI1800-C12 to DNA. There is widespread agreement that polymers with strong DNA-binding capacity do not always guarantee high transfection efficiency.26 There is believed to be an optimal level of binding strength, which should be strong enough to compact DNA outside the cells, but not so strong that DNA can be released efficiently inside the cells.26 Moderate C12 grafting could reduce the DNA-binding strength of bPEI1800-C12 to a level that is beneficial to its transfection efficiency. Using central composite design, bPEI1800-C12 with C12 substitution degree of 19.72 was determined to be the optimal bPEI1800-C12 derivative for DNA delivery (S Liu, unpublished data, 2014).
In comparison with alkylated high-molecular-weight PEI, alkylated LMW PEI underwent a different self-assembly process. It was reported that alkylated bPEI25K could assemble into a liposome-like structure at pH 7.0, and this “polycationic liposomes” phenomenon is thought to be common for hydrophobic derivatives of bPEI25K.28 Due to the huge difference in the chain length between bPEI25K and hydrocarbons, it might be impossible for alkylated bPEI25K to form a micelle-like structure. But in the case of LMW PEI (such as bPEI1800), whose chain length is comparable to that of medium-chain fatty acids and hydrocarbons, the micelle-like assembly of bPEI1800-C12 might occur more easily. Additionally, phospholipid-conjugated bPEI1800 was found to assemble into micelle-like nanoparticles by PEI moiety complexing with DNA, resulting in the inner PEI/DNA core and phospholipid moiety along with specially added helper lipids forming the outer lipid monolayer.18 Taken together, alkylated high-molecular-weight PEI is apt to form a liposome-like structure, and alkylated LMW PEI is likely to assemble into a micelle-like structure, but when LMW PEI was conjugated with amphiphilic moieties (such as phospholipid), the assembly process was different from that of PEI grafted with pure hydrophobic moieties (such as hydrocarbon chains).
Interestingly, we found that the TEM result of bPEI1800-C12/DNA polyplexes (Figure 2B) was quite similar to the assumed structure of C14-CO dimer/DNA complexes24 (Figure S5), and bPEI1800-C12 had a similar molecular structure to gemini/twinned surfactant, as illustrated in Figure S5. Thus, bPEI1800-C12 could be seen as an analog of gemini surfactant with bPEI1800 moiety representing the charged/polar head groups with an inherent inner spacer, and C12 chains being the hydrophobic tails, as illustrated in Figure 2C. In accordance with gemini/twinned surfactant, the assembly process of bPEI1800-C12/DNA complexes was also driven by dual forces: 1) the outside, electrostatic interaction between bPEI1800 moiety and DNA; and 2) the inside, cooperative hydrophobic assembly of C12 chains. During the assembly process of bPEI1800-C12/DNA complexes, DNA undergoes coil–globule transition, forming hydrophobic domains along the DNA chain (Figure 2C) in the same manner as cationic surfactant.24,39 To our knowledge, no such analogy or assembly process of alkylated LMW PEI has been reported in the literature so far.
Stability of bPEI/DNA polyplexes is of prime importance for efficient in vivo gene delivery. bPEI1800-C12/DNA polyplexes were found to be quite robust against anion displacement (Figure 3A) and exhibited high in vitro transfection efficiency in the presence of serum (Figure 5). The molecular formula of bPEI1800-C12 is (C2H4N)42(C12H25)42n, where n is the C12 substitution degree,28 so it was thought there would be 8.3 C12 chains in every molecule of bPEI1800-C12-19.72. It has been reported that cationic surfactant with more than one protonation center and hydrophobic chain showed much higher stability than their corresponding monovalent (single hydrophobic chain and single head group).24,26 As such, the high stability of bPEI1800-C12/DNA polyplexes was likely due to the same “chelate effect”26 as gemini surfactant: multi-protonation centers (DNA compacted more tightly) and multiple hydrophobic chains that could assemble into a highly organized micelle-like structure. Moreover, as depicted in Figure 2C, when complexing DNA with bPEI1800-C12, the individual duplex DNA exhibited a coil–globule transition and formed multi-hydrophobic domains24 along the DNA chain, thus the DNA chain was likely “fixed” to the micelle-like particles. This might be another reason for the high stability of bPEI1800-C12/DNA polyplexes.
As shown in Figure 6C and D, most of the bPEI1800-C12/DNA polyplexes accumulated in reticuloendothelial system organs, but it should be noted that they still exhibited efficient gene expression in these reticuloendothelial system organs after systemic delivery. Moreover, it was surprising that luciferase gene expression could be detected even in the head, suggesting the high in vivo stability of these bPEI1800-C12/DNA polyplexes. Given that the bPEI1800-C12/DNA polyplexes developed in this study were rather simple, in order to improve their in vivo biodistribution, one could fabricate functional nanoparticles based on these bPEI1800-C12 derivatives, such as further conjugating with PEG18 or targeting peptide moieties (cRGD, NGR, etc), and one could also develop co-delivery platforms, such as solid lipid-PEI hybrid nanocarrier.19 In particular, the innate brain accumulation property of bPEI1800-C12 could be utilized to develop brain-targeting nanoparticles with further modification. The interesting phenomenon, that bPEI1800-C12/DNA polyplexes partly accumulated in the head/brain after systemic injection, was likely because bPEI1800-C12, being an analog of gemini surfactant, possessed surfactant activity to help passage through the blood–brain barrier to enter the brain, but further work is needed to prove this assumption and to elucidate why bPEI1800-C12/DNA polyplexes accumulated in the scalp rather than the skull.
Conclusion
We successfully grafted bPEI1800 with lauryl chains (C12) and obtained bPEI1800-C12 which could assemble into a micelle-like structure similarly to gemini surfactant. After C12 grafting, stability and cellular uptake of bPEI1800-C12/DNA polyplexes were greatly improved, and bPEI1800-C12/DNA polyplexes showed superior lysosome escape ability, all leading to high in vitro and in vivo transfection efficiency. In the future, we will focus on the fabrication of nanoparticles with favorable in vivo behaviors based on bPEI1800-C12 derivatives. More studies are required to elucidate the brain (head) accumulation property of bPEI1800-C12. The results of this work suggest that hydrophobic modification of LMW bPEI may give rise to a kind of gemini surfactant-like bPEI derivative with totally altered properties and fantastic benefits.
Acknowledgments
This work was financially supported by Beijing Natural Science Foundation (7142114, 2141004), National Natural Science Foundation of China (NSFC 81373342), Youth Foundation of Peking Union Medical College (2012G07), and Innovation Fund for Postgraduates of Peking Union Medical College (No. 2012-1007-017).
Disclosure
The authors report no conflicts of interest in this work.
References
O’Connor L, Glynn B. Recent advances in the development of nucleic acid diagnostics. Expert Rev Med Devices. 2010;7(4):529–539. | ||
Lai WF. In vivo nucleic acid delivery with PEI and its derivatives: current status and perspectives. Expert Rev Med Devices. 2011;8(2): 173–185. | ||
Singha K, Namgung R, Kim WJ. Polymers in small-interfering RNA delivery. Nucleic Acid Ther. 2011;21(3):133–147. | ||
Chumakova OV, Liopo AV, Andreev VG, et al. Composition of PLGA and PEI/DNA nanoparticles improves ultrasound-mediated gene delivery in solid tumors in vivo. Cancer Lett. 2008;261(2):215–225. | ||
Chen D, Liu Y, Lehtinen A, et al. A novel nonviral gene delivery vector: low-molecular-weight polyethylenimine-graft-ovalbumin. J Appl Polym Sci. 2009;114(6):3744–3750. | ||
Jeon O, Yang HS, Lee TJ, Kim BS. Heparin-conjugated polyethylenimine for gene delivery. J Control Release. 2008;132(3):236–242. | ||
Jiang G, Park K, Kim J, Kim KS, Hahn SK. Target specific intracellular delivery of siRNA/PEI-HA complex by receptor mediated endocytosis. Mol Pharm. 2009;6(3):727–737. | ||
Merkel OM, Beyerle A, Librizzi D, et al. Nonviral siRNA delivery to the lung: investigation of PEG-PEI polyplexes and their in vivo performance. Mol Pharm. 2009;6(4):1246–1260. | ||
Günther M, Lipka J, Malek A, Gutsch D, Kreyling W, Aigner A. Polyethylenimines for RNAi-mediated gene targeting in vivo and siRNA delivery to the lung. Eur J Pharm Biopharm. 2011;77(3):438–449. | ||
Aravindan L, Bicknell KA, Brooks G, Khutoryanskiy VV, Williams AC. Effect of acyl chain length on transfection efficiency and toxicity of polyethylenimine. Int J Pharm. 2009;378(1–2):201–210. | ||
Philipp A, Zhao X, Tarcha P, Wagner E, Zintchenko A. Hydrophobically modified oligoethylenimines as highly efficient transfection agents for siRNA delivery. Bioconjug Chem. 2009;20(11):2055–2061. | ||
Hunter AC, Moghimi SM. Therapeutic synthetic polymers: a game of Russian roulette? Drug Discov Today. 2002;7(19):998–1001. | ||
Thomas M, Klibanov AM. Enhancing polyethylenimine’s delivery of plasmid DNA into mammalian cells. Proc Natl Acad Sci U S A. 2002;99(23):14640–14645. | ||
Kim WJ, Chang CW, Lee M, Kim SW. Efficient siRNA delivery using water soluble lipopolymer for anti-angiogenic gene therapy. J Control Release. 2007;118(3):357–363. | ||
Navarro G, Sawant RR, Biswas S, Essex S, Tros de Ilarduya C, Torchilin VP. P-glycoprotein silencing with siRNA delivered by DOPE-modified PEI overcomes doxorubicin resistance in breast cancer cells. Nanomedicine (Lond). 2012;7(1):65–78. | ||
Navarro G, Sawant RR, Essex S, Tros de Ilarduya C, Torchilin VP. Phospholipid-polyethylenimine conjugate-based micelle-like nanoparticles for siRNA delivery. Drug Deliv Transl Res. 2011;1(1):25–33. | ||
Sawant RR, Sriraman SK, Navarro G, Biswas S, Dalvi RA, Torchilin VP. Polyethyleneimine-lipid conjugate-based pH-sensitive micellar carrier for gene delivery. Biomaterials. 2012;33(15):3942–3951. | ||
Ko YT, Kale A, Hartner WC, Papahadjopoulos-Sternberg B, Torchilin VP. Self-assembling micelle-like nanoparticles based on phospholipid-polyethyleneimine conjugates for systemic gene delivery. J Control Release. 2009;133(2):132–138. | ||
Xue HY, Wong HL. Solid lipid-PEI hybrid nanocarrier: an integrated approach to provide extended, targeted, and safer siRNA therapy of prostate cancer in an all-in-one manner. ACS Nano. 2011;5(9):7034–7047. | ||
Zhou J, Yockman JW, Kim SW, Kern SE. Intracellular kinetics of non-viral gene delivery using polyethylenimine carriers. Pharm Res. 2007;24(6):1079–1087. | ||
Won YY, Sharma R, Konieczny SF. Missing pieces in understanding the intracellular trafficking of polycation/DNA complexes. J Control Release. 2009;139(2):88–93. | ||
Moghimi SM, Hunter AC, Murray JC, Szewczyk A. Cellular distribution of nonionic micelles. Science. 2004;303(5658):626–628. | ||
Hunter AC, Moghimi SM. Cationic carriers of genetic material and cell death: a mitochondrial tale. Biochim Biophys Acta. 2010;1797(6–7):1203–1209. | ||
Lleres D, Dauty E, Behr JP, Mély Y, Duportail G. DNA condensation by an oxidizable cationic detergent. Interactions with lipid vesicles. Chem Phys Lipids. 2001;111(1):59–71. | ||
Mohammed-Saeid W, Michel D, El-Aneed A, Verrall RE, Low NH, Badea I. Development of lyophilized gemini surfactant-based gene delivery systems: influence of lyophilization on the structure, activity and stability of the lipoplexes. J Pharm Pharm Sci. 2012;15(4):548–567. | ||
Kirby AJ, Camilleri P, Engberts JB, et al. Gemini surfactants: new synthetic vectors for gene transfection. Angew Chem Int Ed Engl. 2003;42(13):1448–1457. | ||
Navarro G, Essex S, Sawant RR, et al. Phospholipid-modified polyethylenimine-based nanopreparations for siRNA-mediated gene silencing: implications for transfection and the role of lipid components. Nanomedicine. 2014;10(2):411–419. | ||
Masotti A, Moretti F, Mancini F, et al. Physicochemical and biological study of selected hydrophobic polyethylenimine-based polycationic liposomes and their complexes with DNA. Bioorg Med Chem. 2007;15(3):1504–1515. | ||
Lee SY, Huh MS, Lee S, et al. Stability and cellular uptake of polymerized siRNA (poly-siRNA)/polyethylenimine (PEI) complexes for efficient gene silencing. J Control Release. 2010;141(3):339–346. | ||
Thomas M, Lu JJ, Ge Q, Zhang C, Chen J, Klibanov AM. Full deacylation of polyethylenimine dramatically boosts its gene delivery efficiency and specificity to mouse lung. Proc Natl Acad Sci U S A. 2005;102(16):5679–5684. | ||
Yang F, Huang W, Li Y, et al. Anti-tumor effects in mice induced by survivin-targeted siRNA delivered through polysaccharide nanoparticles. Biomaterials. 2013;34(22):5689–5699. | ||
Díaz-Moscoso A, Vercauteren D, Rejman J, et al. Insights in cellular uptake mechanisms of pDNA-polycationic amphiphilic cyclodextrin nanoparticles (CDplexes). J Control Release. 2010;143(3):318–325. | ||
Witte KN, Kim S, Won YY. Self-consistent field theory study of the effect of grafting density on the height of a weak polyelectrolyte brush. J Phys Chem B. 2009;113(32):11076–11084. | ||
Takahashi M, Yoshikawa K, Vasilevskaya VV, Khokhlov AR. Discrete coil–globule transition of single duplex DNAs induced by polyamines. J Phys Chem B. 1997;101(45):9396–9401. | ||
Kichler A, Leborgne C, Coeytaux E, Danos O. Polyethylenimine-mediated gene delivery: a mechanistic study. J Gene Med. 2001;3(2):135–144. | ||
Sahay G, Alakhova DY, Kabanov AV. Endocytosis of nanomedicines. J Control Release. 2010;145(3):182–195. | ||
dos Santos T, Varela J, Lynch I, Salvati A, Dawson KA. Effects of transport inhibitors on the cellular uptake of carboxylated polystyrene nanoparticles in different cell lines. PloS One. 2011;6(9):e24438. | ||
Godbey WT, Barry MA, Saggau P, Wu KK, Mikos AG. Poly(ethylenimine)-mediated transfection: a new paradigm for gene delivery. J Biomed Mater Res. 2000;51(3):321–328. | ||
Mel’nikov SM, Sergeyev VG, Yoshikawa K. Discrete coil–globule transition of large DNA induced by cationic surfactant. J Am Chem Soc. 1995;117:2401–2408. |
Supplementary materials
  | Figure S5 Chemical structures of CTAB; reduced ornithinyl-cysteinyl-tetradecylamide (C14-CO monomer); oxidized ornithinyl-cysteinyl-tetradecylamide (C14-CO dimer); bPEI1800-C12; gemini/twinned surfactant; and C14-CO dimer/DNA complexes. |
Reference
Lleres D, Dauty E, Behr JP, Mély Y, Duportail G. DNA condensation by an oxidizable cationic detergent. Interactions with lipid vesicles. Chem Phys Lipids. 2001;111(1):59–71. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.