Back to Journals » Cancer Management and Research » Volume 13
High Expression of PKM2 Was Associated with the Poor Prognosis of Acute Leukemia
Authors Huang Y, Chen L, Xie J, Han H, Zhu B, Wang L, Wang W
Received 25 July 2021
Accepted for publication 4 October 2021
Published 13 October 2021 Volume 2021:13 Pages 7851—7858
DOI https://doi.org/10.2147/CMAR.S331076
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Yunxiu Huang,1 Lin-mu Chen,2 Jin-ye Xie,1 Hui Han,1 Bao-fang Zhu,1 Luo-jia Wang,1 Wei-jia Wang1
1Department of Laboratory Medicine, Sun Yat-sen University Affiliated Zhongshan Hospital, Zhongshan, Guangdong Province, People’s Republic of China; 2Department of Pharmacy, Sun Yat-sen University Affiliated Zhongshan Hospital, Zhongshan, Guangdong Province, People’s Republic of China
Correspondence: Wei-jia Wang
Sun Yat-sen University Affiliated Zhongshan Hospital, 2 East of Sun Wen Road, Shi Qi District, Zhongshan, 528403, Guangdong Province, People’s Republic of China
Tel +86-76089880354
Email [email protected]
Purpose: To explore the clinical significance of plasma pyruvate kinase M2 (PKM2) in assessing the incidence and prognosis of acute leukemia.
Methods: Plasma samples from 56 acute myeloid leukemia (AML) patients, 40 acute lymphoblastic leukemia (ALL) patients, and 66 plasma samples from healthy individuals were collected. The level of plasma PKM2 was detected by enzyme-linked immunosorbent assay. The clinical significance of PKM2 in acute leukemia was assessed by analyzing receiver operating characteristic and survival curves.
Results: The plasma levels of PKM2 in AML or ALL patients were significantly higher than those in healthy individuals, respectively. PKM2 can be used as a potential diagnostic index with the AUC of 0.827 for AML and 0.837 for ALL. The level of plasma PKM2 in ALL patients with a BCR/ABL-positive genotype was significantly higher than that in patients with a BCR/ABL-negative genotype (p< 0.05). The event-free survival and the overall survival of acute leukemia patients with higher PKM2 expression was worse than those with lower PKM2 expression.
Conclusion: This study showed that higher levels of PKM2 was negatively correlated with the prognosis of acute leukemia. Therefore, PKM2 can be used as a potential index to assess the incidence and prognosis of acute leukemia.
Keywords: acute leukemia, acute myeloid leukemia, acute lymphoblastic leukemia, pyruvate kinase M2
Introduction
Leukemia is a malignant clonal disease with abnormal hematopoietic stem cells, which stagnates in different stages of cell development due to uncontrolled proliferation, disordered differentiation, and blocked apoptosis.1 It can be divided into acute leukemia (AL) and chronic leukemia (CL) according to the degree of cell differentiation and maturation and the natural course of the disease.2 The incidence of AL is higher than CL in China.3 According to the hematopoietic cell lineage involved, AL can be divided into acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML).4 As the gold standard for the diagnosis of leukemia, bone marrow puncture contributes to the differential diagnosis of leukemia, aplastic anemia, myelodysplastic syndrome, immune thrombocytopenia, different types of anemia, and other blood diseases.5 Although bone marrow puncture is a well-established technique, it is still difficult for the patient to accept, especially for children, in clinical practice. There is also a risk of infection at the puncture site, pain, and osteomyelitis.6 Therefore, it is very important to identify other effective auxiliary less-invasive diagnostic indicators.
Pyruvate kinase M2 (PKM2), which converts phosphoenolpyruvate into pyruvate during glycolysis, is a key enzyme for glycolysis regulation and a newly discovered tumor marker.7 There are four isomeric tissue-specific forms of pyruvate kinase in mammals: PKL, PKR, PKM1, and PKM2.8 PKM1 and PKM2 are alternatively spliced into by the single mRNA transcript of the PKM gene.9 PKM2 exists in the form of high-activity tetramer and low-activity dimer.10 Specially, PKM2 in the dimer form regulates the rate-limiting step of glycolysis, which transfers glucose metabolism from the normal respiratory chain to lactic acid production in tumor cells.11 In addition to its role as a metabolic regulator, PKM2 can also act as a protein kinase, prone to tumorigenesis.12,13 Several studies have shown that post-translational modification or changes in expression of PKM2 can significantly affect the proliferation and differentiation of leukemia cells. Wang et al found that interference with PKM2 expression can inhibit the proliferation of human leukemia cells in vitro, arrest the cell cycle in G1 phase and induce apoptosis, which suggests that PKM2 may promote the development of leukemia by promoting proliferation and inhibiting apoptosis of leukemia cells.14 Replacement of wild-type PKM2 with a sumoylation-deficient mutant (K270R) abrogates the interaction with RUNX1, and blocks myeloid differentiation in vitro and in xenograft models.15 The above studies have shown that PKM2 has an effect on the proliferation and differentiation of leukemia cells, but further clinical trials are needed to evaluate the application value of PKM2 in the clinic.
Therefore, the potential clinical value of determining plasma PKM2 levels for diagnosis and prognosis of AL was explored through ELISA, receiver operating characteristic (ROC) curve and survival analysis. Thus, the clinical relevance of PKM2 for diagnosis and prognosis of AL was explored, so as to further optimize the individualized treatment of the disease and improve the survival rate of AL patients.
Materials and Methods
Subjects
AML and ALL patients diagnosed at the Sun Yat-sen University affiliated Zhongshan Hospital from January 2018 to April 2021 were enrolled retrospectively and were in line with the following inclusion criteria: (1) patients diagnosed with AL for the first time; (2) patients satisfying the diagnostic criteria of AL in accordance with the Diagnostic and Therapeutic Criteria of Hematological Diseases, and diagnosed by myelography, hemogram, and flow cytometry. Exclusion criteria: (1) patients with hepatic and renal insufficiency; (2) patients with malnutrition or diabetes; and (3) patients unable to tolerate chemotherapy or allergic to chemotherapy drugs. A total of 56 cases of AML patients including 37 males and 19 females with an average age of 45 years and 29 cases of healthy individuals including 19 males and 10 females with an average age of 42 years were selected. A further 40 cases were ALL patients including 24 males and 16 females with an average age of 21 years and 37 cases of healthy individuals including 21 males and 16 females with an average age of 23 years old were selected. The differences in sex and age between the two experimental groups and their respective control groups were not statistically significant (P>0.05). The initial treatment options for ALL included VDLD and VDCP et al. The initial treatment regimen for AML varied according to the different subtypes of AML. For instance, in patients with acute promyelocytic leukemia (APL), all-trans retinoic acid and arsenic agents are the mostly used, while treatment of non-APL subtypes relies on anthracycline combined with cytarabine. Treatment with normethoxydaunorubicin (IDA) or daunorubicin (DNR) in combination with cytarabine (ARA-C) or with standard doses of ARA-C in combination with IDA or DNR or mitoxantrone is common. We obtained the patients laboratory test data through our hospital’s computerized test system (Nanfang Huiqiao).
Clinical Data Collection and Follow-Up
Clinical data were collected from AML and ALL patients and control groups (healthy individuals) and included age, sex, glucose (GLU), lactate dehydrogenase (LDH), total bile acid (TBA), cholylglycine (CG), nonesterified fatty acids (NEFA), small dense low-density lipoprotein (sdLDL), apolipoprotein E (apoE), total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), triglycerides (TG), apolipoprotein A1 (apoA1), apolipoprotein B100 (apoB100), lipoprotein a (Lp(a)), white blood cell (WBC), red blood cell (RBC), mean corpuscular volume (MCV), hemoglobin (HGB), platelet (PLT), platelet distribution width (PDW), mean platelet volume (MPV), and the presence of fusion genes of PML/RARA, BCR/ABL, and TEL/AML1 in leukemia patients. Patients were followed from 1 January 2018 to 30 April 2021.This study focused on the event-free survival (EFS) and the overall survival (OS) rates. EFS referred to the time of recurrence or death due to lack of complete remission or the time from diagnosis to the last follow-up. OS was defined as the time from diagnosis to death or the last follow-up.
ELISA
An ELISA kit (Jianglai Biology, Shanghai, China, Cat. #JL48224) was used to measure the concentration of PKM2 in the plasma samples. The optical density (OD) value of each well was measured at a 450 nm wavelength, and a standard curve was drawn to calculate the plasma PKM2 concentration. Required instruments and equipment included the Enzyme labeler (Thermo Fisher, MULTIDKSN FC) and a 37°C incubator (OLABO, HH-W600).
Statistical Methods
GraphPad 8.0 and SPSS 26.0 software were used for statistical analysis of data. The Chi-square test was used for non-parametric data, and mean ± standard deviation (SD) was used for descriptive continuous variables. The independent samples t-test was used for data analysis if the data obeys a normal distribution. If the data did not obey the normal distribution, the Mann–Whitney U-test was used for data analysis. The Kaplan-Meier method was used for the analysis of survival curves, and log-rank method was used for inspection. A p-value<0.05 indicated the difference was statistically significant.
Results
Clinical Characteristics of the Study Participants
The clinical characteristics of AML and ALL patients and of the 66 healthy individuals used in this study are shown in Table 1.
 |
Table 1 Characteristics of the Participants |
Plasma Levels of PKM2 Were Increased in AML and ALL Patients
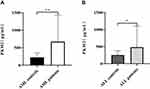
Subhra et al reported that during tumor onset, inflammatory Tumor-associated macrophages (TAMs) induce PKM2 expression, which upon subsequent activation (in its tetrameric form) switches these cells to an immunosuppressive (M2) phenotype in established tumors.16 As shown in Figure 1A, AML patients displayed significantly higher levels of PKM2 (593.11±495.30 pg/mL) compared with healthy individuals (268.12±131.67 pg/mL, p=0.005). Furthermore, the expression of PKM2 in the plasma in the ALL patients (495.05±327.27 pg/mL) was significantly higher compared with healthy individuals (230.13±147.63 pg/mL, p=0.03) (Figure 1B).
Diagnostic Value of Serum PKM2
We evaluated the diagnostic value of serum PKM2 levels for AML or ALL. A ROC curve (Figure 2) showed that the expression of PKM2 could distinguish AML patients from the healthy individuals, and the AUC was 0.827. Moreover, the sensitivity was 91.2%, and the specificity was 66.7% (Supplementary Table 1). The expression of PKM2 could distinguish ALL patients from the healthy individuals, and the AUC was 0.837. Moreover, the sensitivity was 88.2%, and the specificity was 66.7% (Supplementary Table 1).
Comparison of Plasma PKM2 Levels and the Presence of Common Fusion Genes in Leukemia Patients
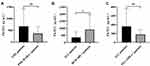
The presence of the PML/RARA fusion gene in AML patients was 23.33%, while that of BCR/ABL and TEL/AML1 fusion genes in ALL patients were 19.56% and 15.22%, respectively. Further statistics showed that the level of PKM2 in patients presenting the BCR/ABL fusion gene was significantly higher than that in patients not expressing this fusion gene (p<0.05). However, the levels of PKM2 in patients harboring the PML/RARA gene fusion and TEL/AML1 gene fusion showed no significant differences compared with other patients (Figure 3).
Survival Analysis of Acute Leukemia Patients
The Kaplan-Meier method was used to construct the survival curves of AL patients stratifying patients according to high- or low-expression of PKM2, and the log-rank method was used for comparisons. According to survival analysis, the EFS and OS of AL patients with high PKM2 expression were significantly lower than those with low PKM2 expression (p<0.05, Figure 4 and Supplementary Table 2). Further, high expression of PKM2 in plasma samples of AL patients was a significant correlated with the poor prognosis of patients.
Discussion
Glycolysis is an important process of tumor metabolism in tumor cells.17 With insufficient oxygen, tumor cells can convert most glucose into lactic acid via glycolysis, thus consuming large amounts of glucose.18 This metabolic pathway characterized by aerobic glycolysis and lactic acid production is called the “Warburg effect”.18 Key enzymes in glycolysis play an important role in the growth, invasion, metastasis, and drug resistance of solid tumors and in AL. By inhibiting glycolysis and its key enzymes, the growth of tumor cells can be inhibited, the apoptosis of tumor cells can be accelerated, and the resistance of tumor cells to treatment can be reversed, so as to achieve effective anti-tumor activity.19
Previous studies have confirmed that PKM2 is a key enzyme regulating glucose metabolism in solid tumors such as breast cancer.20,21 Our study found that PKM2 was also highly expressed in AL, which may be closely related to the vigorous glycolytic metabolic activity characterizing leukemia cells. Increased expression of PKM2 leads to increased glucose uptake, accumulation of glycolytic metabolites, and metabolic reprogramming in cancer cells, which provide advantages for cancer. In addition, metabolic pathways are shifted toward the pentose phosphate pathway (PPP), uronic acid pathway (UAP), and polyol pathway (PYP), leading to an increase in fructose-1,6-diphosphate and non-essential amino acids, thus providing substrates supporting tumor proliferation.22 Conversely, the dimer PKM2 can enter the nucleus as a transcription factor to activate the expression of target genes, and can also coordinate transcription factors targeting other genes to further regulate gene transcription, thus influencing various signaling pathways promoting tumor development.23,24 In addition, the results of survival analysis showed that the time to AL relapse was relatively shorter in patients with high levels of PKM2. Accordingly, AL patients with higher PKM2 expression exhibited shorter OS. Altogether, our findings demonstrated that high expression of PKM2 was associated with the poor prognosis of AL. Furthermore, Sun et al also found that the expression of PKM2 was significantly up-regulated in the retinoid-resistant cell line NB4-R1 of APL, suggesting that PKM2 may be involved in the formation of drug resistance mechanisms of APL cells, thereby affecting the therapeutic outcome and prognosis of patients.25 In addition to the classic metabolic enzyme function in tumor progression, PKM2 can also interfere with the activity of non-metabolic enzymes in tumor regulation, which include metabolic enzymes involved in autophagy.14
The PML-RARA fusion gene is not only a marker for diagnosis and monitoring of minor residual disease (MRD) in AML patients, but also an ideal target for molecular targeted therapy.26 Common fusion genes in ALL patients include the BCR/ABL fusion gene and the TEL/AML1 (ETV6 /RUNX1) fusion gene with an incidence of ~6%–35%, and the latter is considered to be a fusion genes associated with good prognosis.27 Herein, we also found that the expression of PKM2 in ALL patients with BCR/ABL gene fusion was higher than that in other patients without gene fusion, while there was no statistically significant difference in the expression level of PKM2 between AML patients with PML-RARA gene fusion or ALL patients with TEL/AML1 gene fusion. Studies have confirmed that BCR-ABL fusion gene encodes BCR-ABL fusion protein and drives tumor transformation of hematopoietic stem cells, which can stimulate the PI3K/Akt/mTOR signal transduction and lead to impaired autophagy of leukemia cells.28,29 Some researchers have also reported that inhibiting the transcription of BCR-ABL can decrease the expression of PKM2, leading to autophagy and death of leukemia cells,30,31 which is also consistent with the results of this experiment.
In conclusion, this study showed that PKM2 was highly expressed in patients with AL, and the higher the expression level, the worse the prognosis of patients. However, the sample size used in this study was relatively small, and only the expression of PKM2 protein in human peripheral blood was explored. Therefore, a larger sample of clinical patients should be evaluated to further verify our findings. Other factors affecting prognosis of AL disease including, risk stratification, co-expression with fusion genes, gene mutations and their relationship with PKM2 should be analyzed in future studies. Moreover, the mechanisms involving PKM2 in AL, and its regulation of specific signaling pathways should also be explored in subsequent studies.
Abbreviations
PKM2, pyruvate kinase M2 hypotype; AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia; ROC, receiver operating characteristic; EFS, event-free survival; OS, overall survival; GLU, glucose; LDH, lactate dehydrogenase; TBA, total bile acid; CG, cholyglycine; NEFA, nonesterified fatty acids; sdLDL, small dense low-density lipoprotein; apoE, apolipoprotein E; TC, total cholesterol; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TG, triglycerides; apoA1, apolipoprotein A1; apoB100, apolipoprotein B100; Lp(a), lipoprotein a; WBC, white blood cell; RBC, red blood cell; MCV, mean corpuscular volume; HGB, hemoglobin; PLT, platelet; PDW, platelet distribution width; MPV, mean platelet volume.
Ethics Approval
This study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Board of the Sun Yat-sen University affiliated Zhongshan Hospital. This study results had no influence on the subsequent management of patients.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Acknowledgment
We would like to thank all the patients and controls that participated in this study.
Author Contributions
All authors made a significant contribution to the work reported, whether in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas; and took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by the National Nature Science Foundation of China (grant number: 81301492), the Medical Scientific Research Foundation of Guangdong Province of China (grant number: A2021355) and Zhongshan Social Welfare and Foundation Special Project (grant number: K2020B3004).
Disclosure
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
1. Gao J, Wang F, Wu P, Chen Y, Jia Y. Aberrant LncRNA expression in leukemia. J Cancer. 2020;11(14):4284–4296. doi:10.7150/jca.42093
2. Cazzola M. Introduction to a review series: the 2016 revision of the WHO classification of tumors of hematopoietic and lymphoid tissues. Blood. 2016;127(20):2361–2364. doi:10.1182/blood-2016-03-657379
3. Soverini S, Bassan R, Lion T. Treatment and monitoring of Philadelphia chromosome-positive leukemia patients: recent advances and remaining challenges. J Hematol Oncol. 2019;12(1):39. doi:10.1186/s13045-019-0729-2
4. Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. doi:10.1182/blood-2016-03-643544
5. Nguyen TV, Melville A, Nath S, et al. Bone marrow recovery by morphometry during induction chemotherapy for acute lymphoblastic leukemia in children. PLoS One. 2015;10(5):e0126233. doi:10.1371/journal.pone.0126233
6. Bain BJ. Morbidity associated with bone marrow aspiration and trephine biopsy - a review of UK data for 2004. Haematologica. 2006;91(9):1293–1294.
7. Li C, Zhao Z, Zhou Z, Liu R. PKM2 promotes cell survival and invasion under metabolic stress by enhancing Warburg effect in pancreatic ductal adenocarcinoma. Dig Dis Sci. 2016;61(3):767–773. doi:10.1007/s10620-015-3931-2
8. Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43(7):969–980. doi:10.1016/j.biocel.2010.02.005
9. Chen D, Wei L, Liu ZR, et al. Pyruvate kinase M2 increases angiogenesis, neurogenesis, and functional recovery mediated by upregulation of STAT3 and focal adhesion kinase activities after ischemic stroke in adult mice. Neurotherapeutics. 2018;15(3):770–784. doi:10.1007/s13311-018-0635-2
10. Stone OA, El-Brolosy M, Wilhelm K, et al. Loss of pyruvate kinase M2 limits growth and triggers innate immune signaling in endothelial cells. Nat Commun. 2018;9(1):4077. doi:10.1038/s41467-018-06406-8
11. Li YH, Li XF, Liu JT, et al. PKM2, a potential target for regulating cancer. Gene. 2018;668:48–53. doi:10.1016/j.gene.2018.05.038
12. Zahra K, Dey T, Ashish MSP, Pandey U, Pandey U. Pyruvate kinase M2 and cancer: the role of PKM2 in promoting tumorigenesis. Front Oncol. 2020;10:159. doi:10.3389/fonc.2020.00159
13. Zhang Z, Deng X, Liu Y, Liu Y, Sun L, Chen F. PKM2, function and expression and regulation. Cell Biosci. 2019;9(1):52. doi:10.1186/s13578-019-0317-8
14. Wang L, Yang L, Yang Z, et al. Glycolytic enzyme PKM2 mediates autophagic activation to promote cell survival in NPM1-mutated leukemia. Int J Biol Sci. 2019;15(4):882–894. doi:10.7150/ijbs.30290
15. Xia L, Jiang Y, Zhang XH, et al. SUMOylation disassembles the tetrameric pyruvate kinase M2 to block myeloid differentiation of leukemia cells. Cell Death Dis. 2021;12(1):101. doi:10.1038/s41419-021-03400-9
16. Biswas SK. Metabolic reprogramming of immune cells in cancer progression. Immunity. 2015;43(3):435–449. doi:10.1016/j.immuni.2015.09.001
17. Gao P, Shen S, Li X, et al. Dihydroartemisinin inhibits the proliferation of leukemia cells K562 by suppressing PKM2 and GLUT1 mediated aerobic glycolysis. Drug Des Devel Ther. 2020;14:2091–2100. doi:10.2147/DDDT.S248872
18. Lebelo MT, Joubert AM, Visagie MH. Warburg effect and its role in tumourigenesis. Arch Pharm Res. 2019;42(10):833–847. doi:10.1007/s12272-019-01185-2
19. Wu H, Zhao H, Chen L. Deoxyshikonin inhibits viability and glycolysis by suppressing the Akt/mTOR pathway in acute myeloid leukemia cells. Front Oncol. 2020;10:1253. doi:10.3389/fonc.2020.01253
20. Wang Y, Zhao H, Zhao P, Wang X. Targeting PKM2 promotes chemosensitivity of breast cancer cells in vitro and in vivo. Cancer Biomarker. 2021;32(2):221–230. doi:10.3233/CBM-210111
21. Apostolidi M, Vathiotis IA, Muthusamy V, et al. Targeting pyruvate kinase M2 phosphorylation reverses aggressive cancer phenotypes. Cancer Res. 2021;81(16):4346–4359. doi:10.1158/0008-5472.CAN-20-4190
22. Li Z, Yang P, Li Z. The multifaceted regulation and functions of PKM2 in tumor progression. Biochim Biophys Acta. 2014;1846(2):285–296. doi:10.1016/j.bbcan.2014.07.008
23. Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45(5):598–609. doi:10.1016/j.molcel.2012.01.001
24. Wong N, Ojo D, Yan J, Tang D. PKM2 contributes to cancer metabolism. Cancer Lett. 2015;356(2Pt A):184–191. doi:10.1016/j.canlet.2014.01.031
25. Sun ML, Wang GJ, Li J, et al. [Construction of shRNA eukaryotic expression vectors of pkm2 gene and their effect on drug resistant cell line of acute promyelocytic leukemia]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2010;18(1):85–89. Chinese.
26. Cicconi L, Fenaux P, Kantarjian H, Tallman M, Sanz MA, Lo-Coco F. Molecular remission as a therapeutic objective in acute promyelocytic leukemia. Leukemia. 2018;32(8):1671–1678. doi:10.1038/s41375-018-0219-5
27. Organista-Nava J, Gomez-Gomez Y, Illades-Aguiar B, Leyva-Vazquez MA. Regulation of the miRNA expression by TEL/AML1, BCR/ ABL,MLL/AF4 and TCF3/PBX1 oncoproteins in acute lymphoblastic leukemia (Review). Oncol Rep. 2016;36(3):1226–1232. doi:10.3892/or.2016.4948
28. Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5(3):172–183. doi:10.1038/nrc1567
29. Niu Y, Yang X, Chen Y, et al. EVI1 induces autophagy to promote drug resistance via regulation of ATG7 expression in leukemia cells. Carcinogenesis. 2020;41(7):961–971. doi:10.1093/carcin/bgz167
30. Shinohara H, Taniguchi K, Kumazaki M, et al. Anti-cancer fatty-acid derivative induces autophagic cell death through modulation of PKM isoform expression profile mediated by bcr-abl in chronic myeloid leukemia. Cancer Lett. 2015;360(1):28–38. doi:10.1016/j.canlet.2015.01.039
31. De Rosa V, Monti M, Terlizzi C, Fonti R, Del Vecchio S, Iommelli F. Coordinate modulation of glycolytic enzymes and OXPHOS by imatinib in BCR-ABL driven chronic myelogenous leukemia cells. Int J Mol Sci. 2019;20(13):3134. doi:10.3390/ijms20133134
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.