Back to Journals » OncoTargets and Therapy » Volume 10
High expression of long noncoding RNA HULC is a poor predictor of prognosis and regulates cell proliferation in glioma
Authors Yan H, Tian R, Zhang M, Wu J, Ding M, He J
Received 13 October 2016
Accepted for publication 18 November 2016
Published 21 December 2016 Volume 2017:10 Pages 113—120
DOI https://doi.org/10.2147/OTT.S124614
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yao Dai
Hong Yan,* Rui Tian,* Min Zhang, Jing Wu, Min Ding, Jie He
Department of Pathology, Anhui Provincial Hospital affiliated to Anhui Medical University and Anhui Provincial Cancer Hospital, Hefei, Anhui, People’s Republic of China
*These authors contributed equally to this work
Purpose: Emerging studies show that long noncoding RNAs (lncRNAs) have important roles in carcinogenesis. This study investigated the role of lncRNA highly upregulated in liver cancer (HULC) expression in glioma and its clinical significance in glioma patients.
Materials and methods: HULC expression was detected in glioma tissues and cell lines by using real-time quantitative reverse transcription polymerase chain reactions. Association between HULC levels and clinicopathological factors and patients prognosis was also analyzed. Expression of HULC was restored and knocked down in glioma cell line U87 by using HULC cDNA and siRNA, respectively. CCK-8 and colony formation assays were used to investigate the role of HULC in the regulation of proliferation of glioma cells.
Results: HULC was highly expressed in glioma tissues, being closely related to age and grade of glioma. Univariate survival analysis demonstrated that high HULC levels were significantly associated with overall survival (OS) (hazard ratio [HR], 0.422; 95% confidence interval [CI], 0.220–0.806; P=0.009), and it remained an independent predictor for OS (HR, 0.340; 95% CI, 0.175–0.659; P=0.001) in multivariate Cox regression analysis. Functionally, forced expression of HULC results in increased cell proliferation and colony formation of U87 glioma cell line, whereas knockdown of HULC expression reduced these oncogenic properties of glioma cells.
Conclusion: These findings suggest that HULC may play an important role in glioma progression and will be further evaluated as a biomarker for predicting the survival of glioma patients.
Keywords: long noncoding RNA, HULC, glioma, prognostic biomarker
Introduction
Glioma accounts for the great majority of primary tumors in the central nervous systems of adults.1 The prognosis of glioma is poor, with a median survival of only 12–15 months.2 One and five year survival rates are <20% and 5%, respectively.3 Therefore, it is necessary to identify novel biomarkers specific to the stages of glioma or to the susceptibility of gliomas to anticancer agents, which may be helpful to develop more rational and effective therapies.
Long noncoding RNAs (lncRNAs) are transcripts longer than 200 nucleotides with little or no protein coding function. They are considered a new class of regulatory ncRNAs. In recent decades, it has been reported that aberrant expression of many lncRNAs play important roles in a large range of biological processes and diseases, especially in cancer biology.4 Furthermore, lncRNAs are frequently observed in cancers involved in carcinogenesis and cancer progression which indicates that dysregulated lncRNAs could probably serve as novel biomarkers for early diagnosis, effective therapeutic targets, and prognosis prediction of malignant tumors. For instance, overexpression of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) indicates worse prognosis in lung cancer patients.5 LncRNA ZEB1-AS1 promotes tumorigenesis and predicts poor prognosis in glioma.6 Increased lncRNA SNHG20 indicates a poor prognosis for colorectal cancer.7 Together, these observations provide evidence and support for potential roles for lncRNAs in tumor development and progression.
Highly upregulated in liver cancer (HULC), located on chromosome 6p24.3 and conserved in primates, is overexpressed in hepatocellular carcinoma (HCC).8 Aberrant expression of lncRNA HULC has been reported in HCC,9,10 and its potential as a diagnostic biomarker has been proposed.11,12 In addition to its influence in HCC, HULC also plays an important role in the growth and tumorigenesis of human gastric cancer (GC).13 HULC may also serve for regulating growth in human pancreatic cancer.14 However, the implication of HULC in glioma was still unclear. The present study determined the expression level of HULC in glioma tissues and cell lines and then explored its clinical values as a biomarker for diagnosis and prognosis prediction of glioma. Later, it further investigated the role of HULC expression in the regulation of glioma cell proliferation and colony formation in vitro.
Materials and methods
Patients and tissue specimens
Seventy glioma samples were acquired from patients who experienced surgery resection from May 2011 to September 2015 at the Department of Neurosurgery, Anhui Cancer Hospital (Heifei, People’s Republic of China). Twelve normal brain tissues were used as controls. Disease diagnosis and classification were made by pathological examination based on the World Health Organization (WHO) classification system. Tumor and normal specimens were snap-frozen in liquid nitrogen and stored at −80°C immediately after resection. Patients who had ever undergone radiotherapy, chemotherapy, and surgery were excluded from this study. The protocol of this study was approved by the Ethics Committee of Anhui Provincial Hospital affiliated to Anhui Medical University. The work undertaken conforms to the provisions of the Declaration of Helsinki. Written informed consent was obtained from all patients.
Cell culture and transfection
The human glioma SHG44, U251, and U87 cell lines were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA) and were cultured in Dulbecco’s Modified Eagle’s Medium with 10% fetal bovine serum (GIBCO, Carlsbad, CA, USA) in a humidified atmosphere of 5.0% CO2 at 37°C. HULC expression was modified by gene knockdown through lentivirus vector. For knockdown, si-HULC (target sequence: 5′-GCCTTTACAAGGGAATGAAGA-3′) was used for further studies. The pCDNA3.1-HULC plasmid and siRNA were synthesized by GenePharma (Shanghai, China), both using Lipofecamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s directions. Assays were conducted at 48 h after transfection.
RNA isolation and quantitative polymerase chain reaction (PCR)
Total RNA was isolated from tissue samples or cultured cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. cDNA was generated by reverse transcription of 1 μg of total RNA using Thermo Scientific RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Wilmington, DE, USA). Quantitative reverse transcription polymerase chain reaction (qRT-PCRs) were conducted using SYBR Green real-time PCR Kit (Takara, Dalian, China) to detect the expression of lncRNA HULC. Results were normalized to an endogenous control (GAPDH). The primers for HULC were 5′-ACCTCCAGAACTGTGATCCAAAATG-3′ and 5′-TCTTGCTTGATGCTTTGGTCTG-3′; the primers for GAPDH were 5′-CCCATCACCATCTTCCAGGAG-3′ and 5′-GTTGTCATGGATGACCTTGGC-3′.
Cell viability assay
Cell viability was measured with the Cell Counting Kit-8 (CCK-8) assay. Briefly, after exposing U87 cell to HULC overexpression or silencing for 1–4 days, 20 μL of CCK-8 (Beyotime, Shanghai, China) dissolved in phosphate-buffered saline (PBS) at a concentration of 5 mg/mL was added to the cancer cells and incubated in a CO2 incubator for 3 h. Cell viability was determined by measuring the absorbance of the converted dye at 450 nm. All the experiments were carried out in sextuplicate and repeated three times.
Colony formation assay
Both nontransfected and transfected U87 cells were seeded in six-well plates at the density of 1,000 cells/well. After ~9 days, the cells were fixed with 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet (Beyotime Institute of Biotechnology, Shanghai, China) for 30 min. Next, the plates were washed mildly with PBS before being air-dried, and the stained colonies were photographed using a high-resolution camera. The experiments were performed at least in triplicate.
Statistical analyses
Statistical Package for the Social Sciences (SPSS) 19.0 software system (IBM Corporation, Armonk, NY, USA) was used for statistical analysis. All values were described as mean ± standard deviation. The Student’s t-test was used for comparisons between groups. A chi-square test was applied to determine the association of HULC levels with clinicopathological features. Overall survival (OS) was the time interval from the date of surgery to death from glioma or to the most recent follow-up. Survival curves were estimated with the Kaplan–Meier method, and differences were compared using the log-rank test. Univariate and multivariate analyses were performed using Cox proportional hazards regression models, and hazard ratios (HRs) for variables corresponding to OS were calculated. And a two-sided P-value of <0.05 was considered statistically significant.
Results
Expression of lncRNA HULC is upregulated in glioma tissues
To investigate the implication of this lncRNA in glioma, this study detected the expression levels of HULC in 70 glioma tissues and 12 normal brain tissues by qRT-PCR. As shown in Figure 1A and Table 1, expression of HULC was statistically upregulated in glioma tissues compared with the normal brain tissues (both P<0.01).
  | Table 1 Expression of HULC in glioma tissues and normal brain tissues |
Relationship of lncRNA HULC expression with clinicopathological features of glioma patients
To further elucidate the significance of HULC in glioma, we calculated the correlation of HULC expression with clinicopathological features of 70 glioma patients (Table 2), finding that highly expressed HULC was positively associated with age and tumor grade (P=0.024 and P<0.001, respectively). However, no association of HULC expression with gender, tumor size, tumor location, and tumor nodule number was detected. Thus, it was speculated that HULC might have important implications for the progression of glioma.
High expression levels of lncRNA HULC were significantly related to reduced OS of glioma patients
In order to further determine the prognostic significance of lncRNA HULC in glioma, this study examined the relationship of HULC levels with OS rates through Kaplan–Meier analysis and log-rank test in 70 glioma cases, discovering that high HULC expression might predict a poor OS (P=0.007; Figure 1B).
LncRNA HULC expression was a potentially independent prognostic marker for glioma patients
Results of univariate Cox regression analysis revealed that HULC overexpression (HR =0.422, 95% CI: 0.220–0.806, P=0.009), age (HR =0.505, 95% CI: 0.262–0.974, P=0.042), and the clinical grade (HR =0.236, 95% CI: 0.072–0.773, P=0.017) were prime variables for glioma prognosis (Table 3). Furthermore, with gender, tumor size, tumor location, and tumor nodule number as covariates, multivariate Cox regression analysis indicated that highly expressed HULC (HR =0.340, 95% CI: 0.175–0.659, P=0.001) and age (HR =0.390, 95% CI: 0.200–0.762, P=0.006) were independent prognostic factors for OS of glioma prognosis (Table 3).
Manipulation of HULC levels in glioma cancer cells
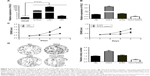
Next, qRT-PCR analysis was performed to examine the expression levels of HULC in glioblastoma cell lines and normal brain tissues, finding that they were significantly higher in three high-degree glioblastoma cell lines (SHG44, U87, U251) than in the normal brain tissues (P<0.01; Figure 2A).
To further examine the role of HULC in glioma, U87 cell line was selected for further analysis, and a pCDNA/HULC vector or si-HULC was transfected into U87 cells, respectively. qRT-PCR analysis of HULC levels was performed at 48 h post-transfection and revealed that HULC expression was increased 118.33-fold in U87 cells, compared with control cells. However, the si-HULC could silence HULC expression effectively in U87 cells, compared with the nonspecific siRNA (si-NC) groups (P<0.01; Figure 2B).
Effect of HULC on cell proliferation in vitro
The significant increase of HULC expression in glioma samples prompted us to explore the possible biological significance of HULC in tumorigenesis. Cell proliferation was examined through CCK-8 and colony formation assays after overexpression or depletion of HULC in U87 cells. CCK-8 assay revealed that cell growth was significantly increased in pCDNA/HULC-transfected cells compared with respective controls (P<0.01; Figure 2C, left), while proliferation was impaired in U87 cells transfected with si-HULC (P<0.01; Figure 2C, right). Similarly, the result of colony formation assay revealed that the number of U87 cell colonies was significantly increased by forced expression of HULC or decreased following inhibition of HULC in U87 cells (Figure 2D).
Discussion
To date, >3,000 lncRNAs have been identified and only a small number of functional lncRNAs have been well characterized.15 Increasing reports of aberrant expression of lncRNAs across numerous cancer types suggested that some lncRNAs could promote the formation and development of tumors.16–18 With regard to these latter effects, expression levels of certain lncRNAs have been shown to be associated with the recurrence, metastasis, and prognosis of various cancers (HCC, gastric carcinoma, and breast carcinoma).19–21 Therefore, lncRNAs may provide a missing piece of the otherwise well-known oncogenic and tumor suppressor network puzzle.
Glioma is a complex genetic disease and despite all the progress achieved by basic and clinical research in recent years, the molecular genetic mechanisms involved in the pathogenesis of glioma are not yet clear. LncRNA expression has not been well explored in glioma, and the knowledge of how lncRNAs act in glioma cells remains very limited. This study evaluated the expression of HULC, which is a lncRNA that is associated with the metastatic processes of tumors in glioma samples.8,13,14,22
Previous studies reported HULC was a negative prognostic factor for liver cancer,8 and clinical data showed that HULC expression in HCC tissues was significantly increased as compared with that observed in normal liver tissues.15,23 Moreover, HULC plays pivotal roles in aberrant lipid metabolism in HCC through miR-9/PPARA/ACSL1/cholesterol/RXRA/HULC signaling.24 In addition, hepatitis B virus X protein (HBx) could upregulate HULC expression in human immortalized normal liver LO2 cells and hepatoma HepG2 cells, and upregulation of HULC by HBx could promote proliferation of hepatoma cells through suppressing p18.25 Based on the previous reports, HULC plays an important role in liver carcinogenesis and acts as an oncogenic ncRNA. HULC has been demonstrated to act as a microRNA (miRNA) sponge that binds to and reduces the expression of a number of miRNAs, including miR-372. The reduction of miR-372 expression leads to increased expression of its target PRKACB, which in turn induces CREB phosphorylation. This process results in alterations in the patterns of the deacetylation and methylation of histones, which leads to chromatin remodeling and influences the expression of a series of genes.26
This study first discovered that the expression of lncRNA HULC significantly increased in glioma tissues. Furthermore, it explored the association between HULC expression and clinical characteristics of glioma patients, finding that its expression was significantly related with age and tumor grade, suggesting its crucial role in glioma. Analyses of the relationship between high versus low levels of HULC expression in glioma patient survival, as performed using the Kaplan–Meier analysis and log-rank test, indicated that overexpression of HULC had a decreased rate of OS. More importantly, multivariate Cox regression analysis provided evidence that both age and HULC expression could serve as independent poor prognostic factors in patients with glioma. These results are in accord with other findings which show that HULC is correlated with tumor size, lymph node metastasis, and vascular invasion in gastric and pancreatic cancers.13,14
In addition, research on function and mechanism of HULC was conducted in cell models to determine the precise role of HULC in glioma. Sun et al provided evidence that highly expressed HULC could enhance the proliferation, migration, and invasion ability of osteosarcoma cells.27 This study observed that elevated expression of HULC promoted proliferation and colony formation of glioma cells, which HULC depletion inhibited cell viability and colony formation in vitro. These findings revealed that HULC might be involved in glioma progression, stimulating new research directions and therapeutic options. Li et al discovered that high expression of HULC, greatly inducing the expression and activity of HULC, which were associated with an increase in epithelial-mesenchymal transition (EMT)-related parameters by sequestering miR-200a-3p in HCC cells.23 Thus, it is speculated that lncRNA HULC could affect the cell migration and invasion through the HULC–EMT pathway in glioma patients. Liu et al indicated that the competitive endogenous RNA (ceRNA) activity of lncRNA HOX transcript antisense intergenic RNA (HOTAIR) imparted a miRNA/lncRNA trans-regulatory function to protein-coding mRNAs, and there may be many other lncRNAs that function as ceRNAs to regulate the expression of key genes in cancer.28 Thus, the identification of ceRNAs will undoubtedly enhance the knowledge of how lncRNAs function, allowing us to better understand the pathogenesis and development of cancer and ultimately facilitate the development of lncRNA-directed diagnostics and therapeutics against this deadly disease.
Conclusion
In conclusion, this research confirmed elevated lncRNA HULC expression in glioma tissues and cell lines, and its overexpression may be indicative of higher risks for glioma progression and poor OS rate. These findings suggested that HULC would be an underlying prognostic marker and a new potential therapeutic target for the treatment of human gliomas. Manipulation of HULC expression using the gain and loss of function approaches affected glioma cell viability, colony formation, and gene expression in vitro. Future studies will further investigate the underlying molecular mechanisms by which HULC is overexpressed in glioma, and HULC expression contributes to glioma progression.
Acknowledgment
This study was supported by grants from The National Natural Science Foundation of China (Nos 81502323 and 81272800).
Disclosure
The authors report no conflicts of interest in this work.
References
Sathornsumetee S, Rich JN. New treatment strategies for malignant gliomas. Expert Rev Anticancer Ther. 2006;6(7):1087–1104. | ||
Gurley SN, Abidi AH, Allison P, et al. Mechanism of anti-glioma activity and in vivo efficacy of the cannabinoid ligand KM-233. J Neuro Oncol. 2012;110(2):163–177. | ||
Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64(19):6892–6899. | ||
Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. | ||
Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. | ||
Lv QL, Hu L, Chen SH, et al. A long noncoding RNA ZEB1-AS1 promotes tumorigenesis and predicts poor prognosis in glioma. Int J Mol Sci. 2016;17(9):1431. | ||
Li C, Zhou L, He J, Fang XQ, Zhu SW, Xiong MM. Increased long noncoding RNA SNHG20 predicts poor prognosis in colorectal cancer. BMC Cancer. 2016;16:655. | ||
Panzitt K, Tschernatsch MM, Guelly C, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132(1):330–342. | ||
Wong TS, Fau LX, Fau WB, Fau NR, Fau YA, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14(9):2588–2592. | ||
Park NJ, Zhou H, Elashoff D, et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15(17):5473–5477. | ||
Liu CJ, Kao SY, Tu HF, Tsai MM, Chang KW, Lin SC. Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral Dis. 2010;16(4):360–364. | ||
Gorenchtein M, Poh CF, Saini R, Garnis C. MicroRNAs in an oral cancer context – from basic biology to clinical utility. J Dent Res. 2012;91(5):440–446. | ||
Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep. 2014;31(1):358–364. | ||
Peng W, Gao W, Feng J. Long noncoding RNA HULC is a novel biomarker of poor prognosis in patients with pancreatic cancer. Med Oncol. 2014;31(12):346. | ||
Yang Z, Lu Y, Xu Q, Tang B, Park CK, Chen X. HULC and H19 played different roles in overall and disease-free survival from hepatocellular carcinoma after curative hepatectomy: a preliminary analysis from gene expression omnibus. Dis Markers. 2015;2015:1–9. | ||
Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15(1):43. | ||
Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. | ||
Li J, Meng H, Bai Y, Wang K. Regulation of lncRNA and its role in cancer metastasis. Oncol Res. 2016;23(5):205–217. | ||
Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25(24):2573–2578. | ||
Mercer TR, Fau DM, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. | ||
Yu FJ, Zheng JJ, Dong PH, Fan XM. Long non-coding RNAs and hepatocellular carcinoma. Mol Clin Oncol. 2015;3(1):13–17. | ||
Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. BioMed Res Int. 2013;2013(23):136106. | ||
Li SP, Xu HX, Yu Y, et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7(27):42431–42446. | ||
Cui M, Xiao Z, Wang Y, et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015;75(5):846–857. | ||
Du Y, Kong G, You X, et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287(31):26302–26311. | ||
Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucl Acids Res. 2010;38(16):5366–5383. | ||
Sun XH, Yang LB, Geng XL, Wang R, Zhang ZC. Increased expression of lncRNA HULC indicates a poor prognosis and promotes cell metastasis in osteosarcoma. Int J Clin Exp Pathol. 2015;8(3):2994–3000. | ||
Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.