Back to Journals » Cancer Management and Research » Volume 14
High Expression of lncRNA HEIH is Helpful in the Diagnosis of Non-Small Cell Lung Cancer and Predicts Poor Prognosis
Authors He C, Huang D, Yang F, Huang D, Cao Y, Peng J, Luo X
Received 3 June 2021
Accepted for publication 16 December 2021
Published 9 February 2022 Volume 2022:14 Pages 503—514
DOI https://doi.org/10.2147/CMAR.S320965
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Beicheng Sun
Chaowen He, Dongxuan Huang, Fan Yang, Dongsheng Huang, Yahui Cao, Jianfeng Peng, Xiaohua Luo
Department of Pulmonary and Critical Care Medicine, Shenzhen Longhua District Central Hospital, Shenzhen, 518110, Guangdong, People’s Republic of China
Correspondence: Chaowen He, Department of Pulmonary and Critical Care Medicine, Shenzhen Longhua District Central Hospital, No. 187 Guanlan Avenue, Longhua District, Shenzhen, 518110, Guangdong, People’s Republic of China, Tel +86-18123964996, Email [email protected]
Background: This study aims to investigate the expression and clinical value of long non-coding RNA (lncRNA) HEIH in peripheral blood of patients with non-small cell lung cancer (NSCLC).
Methods: Healthy subjects (N=70), patients with lung squamous cell carcinoma (LUSC, N=70) and patients with lung adenocarcinoma (LUAD, N=80) were included. LncRNA HEIH expression in peripheral blood of included subjects was detected using RT-qPCR. According to the median expression of lncRNA HEIH, LUSC and LUAD patients were allocated into lncRNA HEIH high/low expression groups. The correlation between lncRNA HEIH and clinical indicators of patients was analyzed; Logistic multifactor regression was used to analyze the independent risk factors influencing lncRNA HEIH level. Receiver-operating characteristic (ROC) curve was used to evaluate the diagnostic efficacy of lncRNA HEIH and carcinoembryonic antigen (CEA) in LUSC/LUAD patients. MedCalc-Comparison of ROC curves was used to compare the area under ROC curve. The cumulative survival rates of lncRNA HEIH high/low expression group were analyzed by Kaplan–Meier curve. COX multivariate analysis was used to assess the independent factors affecting prognosis of NSCLC.
Results: LncRNA HEIH in peripheral blood of LUSC/LUAD patients was higher than that in healthy controls, with no evident difference between LUSC and LUAD groups. In LUSC/LUAD patients, TNM stage, lymph node metastasis, distal metastasis, and CEA were independent risk factors affecting lncRNA HEIH; patients with high lncRNA HEIH expression had larger pack-years and tumor size, higher CEA level and tumor stage, and higher risk of lymph node metastasis and distal metastasis. LncRNA HEIH had higher diagnostic efficiency than CEA in NSCLC patients. High expression of lncRNA HEIH predicted poor prognosis in patients with NSCLC and was an independent risk factor for prognosis of NSCLC.
Conclusion: High expression of lncRNA HEIH is helpful in the diagnosis of NSCLC and predicts poor prognosis.
Keywords: non-small cell lung cancer, long non-coding RNA HEIH, prognosis, carcinoembryonic antigen, lung squamous cell carcinoma, lung adenocarcinoma, peripheral blood
Introduction
According to the data from China Cancer Registry Center, the incidence and mortality of lung cancer rank the first among malignant tumors in the world,1 and about 2 million patients die of lung cancer every year,2 among which non-small cell lung cancer (NSCLC) accounted for 80%~85%.3 NSCLC can be divided into lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUCC) and large cell carcinoma (LCC) according to its pathological features. LUAD and LUSC accounted for approximately 50% and 40% of NSCLC patients, respectively.4 LUAD is more common in women and smokers and is the main pathological subtype of lung tumors; there are no special symptoms in the early stage, mainly manifesting as respiratory diseases, including cough, low fever, and chest pain.5,6 LUSC is more common in older men and has historically been strongly associated with smoking, and the main symptoms are fever, cough, hemoptysis, and chest pain.7 In addition, genes in LUSC and LUAD patients were expressed differently.8 For example, the diagnostic marker CEP55 was significantly differentially expressed in LUSC and LUAD.9 Due to the atypical early clinical manifestations of the disease and the lack of biological markers for early diagnosis, about 75% of the patients were already in the middle and advanced stage when detected, and the tumor cells had lymphatic metastasis and distant metastasis, thus losing the best opportunity for surgical treatment and resulting in poor prognosis.10 Therefore, it is of great significance to study the mechanism of the occurrence and development of NSCLC and to obtain novel biomarkers that can be used for early diagnosis, prognosis evaluation and treatment of NSCLC.
Long non-coding RNA (lncRNA) is a class of RNA molecules with a transcript length of more than 200 nt without coding protein function, which can regulate gene expression level, post-transcriptional modification, binding to transcription factors or miRNAs, and play a regulatory role in many biological processes.11 A study has discovered that lncRNAs regulate cell proliferation, growth, and apoptosis, and the abnormal expression of lncRNA is closely linked to the occurrence of tumor.12 More and more evidence indicates the involvement of lncRNAs in the pathogenesis and development of NSCLC.13,14 For instance, lncRNA FeZF1-AS1 is up-regulated in lung cancer and promotes NSCLC through the ITGA11/miR-516b-5p axis;15 LncRNA LCAT1, as an oncogene, can inhibit the growth of lung cancer cells and inhibit the tumorigenesis and metastasis of xenograft mice after knockout.16 Additionally, lncRNA PCAT6 is highly expressed in NSCLC as an oncogene, and knockdown of PCAT6 inhibits NSCLC cell growth by inducing cell cycle arrest and apoptosis at the G1 phase.17 At present, several lncRNAs18 that can be used as candidate tumor biomarkers have been detected in the body fluids of patients, and their research as NSCLC specific biomarkers has been widely reported.19–21 High expression in hepatocellular carcinoma (HEIH) is a lncRNA originally found in HBV-induced hepatocellular carcinoma.22 In patients with hepatocellular carcinoma, high expression of HEIH is associated with an increased risk of recurrence and declined overall survival after surgery. Recent studies have demonstrated that HEIH is also highly expressed in other types of cancers including colorectal cancer, melanoma and NSCLC.23–25 Jia et al found that HEIH was markedly overexpressed in NSCLC tissues and cell lines, which promoted the proliferation and metastasis of NSCLC cells.24 However, the expression level of HEIH in peripheral blood of NSCLC and its clinical value in the diagnosis and prognosis of NSCLC have not been reported yet. This study herein investigated the expression level of lncRNA HEIH in peripheral blood of NSCLC patients and explored its clinical value in the diagnosis and prognosis of NSCLC.
Materials and Methods
Ethics Statement
The recruitment procedure was in accordance with the principles of the Declaration of Helsinki of the World Medical Association. All subjects signed a written informed consent. The study protocol was approved by the ethics committee of Shenzhen Longhua district central hospital (AF/SC-08/01.0).
Study Subjects
We used the software Gpower to pre-estimate the samples. Briefly, we took the effect value as the median value of the system recommendation (effect size d = 0.5), set the parameter α = 0.05, and set the statistical efficacy 1 − β = 0.9. P value was obtained from the bilateral test, and the sample ratio of normal group/disease group = 1/2. The calculated sample size was as follows: normal group ≥ 64, and the NSCLC group ≥ 128. Considering the sample loss (10–15%) and the grouping of NSCLC (LUSC and LUAD groups), we finally obtained a sample size of 70 cases in the normal group and 150 cases in the NSCLC group (LUSC: 70 cases, LUAD: 80 cases) (Supplementary Figure 1). Gpower software was used to calculate the statistical power of the lncRNA HEIH expression difference between the normal group and NSCLC group. According to the effect size d = mean difference/mean standard variance, we calculated that effect size d =1.91. Meanwhile, we set the parameter α=0.05, the sample size was 70 and 150 and calculated the statistical efficacy as 1-β > 0.95 (Supplementary Figure 2), indicating that the selected sample size was statistically significant.
Based on the Gpower analysis, 150 patients with NSCLC admitted to the Department of Respiratory Medicine of Shenzhen Longhua district central hospital from December 2013 to December 2015 were selected as the study subjects. According to the 2004 World Health Organization (WHO) classification of lung tumors,26 NSCLC patients were further divided into 80 lung adenocarcinoma (LUAD) patients and 70 lung squamous cell carcinoma (LUSC) patients. Inclusion criteria for NSCLC were as follows: (a) Having typical clinical manifestations of lung cancer; (b) All cases were diagnosed by histopathology or cytology; and (c) All cases were new and had not received surgery, radiotherapy, chemotherapy or targeted therapy. Exclusion criteria were as follows: (a) A history of tuberculosis; (b) Patients with diabetes; (c) Accompanied by hypertension, hyperlipidemia or hyperglycemia; (d) Suffering from other malignant tumors; (e) Prolonged use of immunosuppressant and steroid hormones; (f) Failure to follow up regularly; and (g) Pregnant or lactating women. Meanwhile, 70 healthy volunteers who came for physical examination at the same period were selected as the control group.
Data and Sample Collection
The age (≤60; >60), gender, smoking, pack-years (< 20; ≥20), comorbidities, and other baseline clinical data of enrolled subjects were recorded, as well as tumor size (≤3 cm; >3 cm), tumor-node-metastasis (TNM) stage (I; II/III), lymph node metastasis (Absent; Present), distal metastasis and carcinoembryonic antigen (CEA) (μg/mL). Among them, comorbidities refer to patients complicated with diseases in the heart, cerebrovascular, respiratory and urinary system or dysfunction. The cumulative illness rating scale for geriatrics was adopted to score the comorbidities of 14 organ systems. According to the severity of disease, the disease score on each scale was rated on a scale of 0–4 points (0: no disease; 1: the current mild disease or past serious disease; 2: moderate dysfunction or disease, requiring first-line treatment; 3: serious/persistent aboriginal diseases; 4: extremely severe damage or failure of the function that needs immediate treatment). The severity index (SI) was used as the evaluation index of comorbidities (SI = total score/number of comorbidities). According to SI, the comorbidities were classified into 3 levels: no comorbidities (SI = 0), mild comorbidities (SI ≤ 2), and severe comorbidities (SI > 2). A total of 2 mL fasting peripheral blood was collected from the vein of all patients without preoperative chemotherapy or radiotherapy, and centrifuged at 4°C and 2000 g for 10 min, and the supernatant was transferred to an EP tube and stored at −80°C until determination.
Enzyme-Linked Immunosorbent Assay (ELISA)
Human CEA levels in peripheral blood of NSCLC patients and healthy subjects were detected using human CEA ELISA Kit (Amyjet Scientific., Wuhan, China).
Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
Trizol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA. The total RNA was transcribed into cDNA by Primescript RT reagent kit (Takara, Dalian, China) and the qPCR assay was performed on the ABI7900HT Fast PCR Real-Time System (Applied Biosystems, Foster city, CA, USA) using SYBR® Prepremix Ex TaqTM II (Takara, Dalian, China). The reaction conditions included pre-denaturation at 95°C for 10 min, and 40 cycles of denaturation at 95°C for 10s, annealing at 60°C for 20s, and extension at 72°C for 34s. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal reference, and the data were analyzed by the 2−ΔΔCT method.27 The primers were synthesized by Sangon Bioengineering Shanghai Co., Ltd (Shanghai, China), and the sequences are shown in Table 1.
 |
Table 1 Primer Sequence |
Statistical Analysis
Statistical software SPSS 21.0 (IBM Corp. Armonk, NY, USA), GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA) and Medcalc® version 15.0 (Medcalc Software Ltd, Ostend, Belgium) were used for data analysis and map plotting. Gpower was used for sample size pre-estimation. Shapiro–Wilk (W-test) test showed that data of numerical variables were normally distributed and were expressed as mean ± standard deviation. Unpaired t-test was used for comparison between two groups, one-way analysis of variance (ANOVA) was used for comparison among multiple groups, and Tukey’s multiple comparisons test was used for post hoc test. Fisher’s exact test was used for comparative analysis of classification variables. Receiver operating characteristic curve (ROC) was used to analyze the diagnostic value of lncRNA HEIH for NSCLC. Kaplan-Meier method was used to analyze the effect of lncRNA HEIH on the prognosis of NSCLC patients. Logistic multifactor regression analysis was used to evaluate the influencing factors of lncRNA HEIH expression. COX regression test was used to analyze independent prognostic factors of NSCLC. The area difference under the ROC curve was analyzed by MedCalc-comparison of ROC curves. A value of P < 0.05 was indicative of statistically significant.
Results
Comparison of Clinicopathological Features Between Patients with NSCLC and Healthy Subjects
A total of 220 subjects were included in this study, including 70 healthy subjects, 70 LUSC patients, and 80 LUAD patients. The comparative analysis of the clinical data of LUSC and LUAD patients and healthy subjects manifested that there were no significant differences in age, gender and smoking among the three groups (P > 0.05), while the pack-years, comorbidities, and serum CEA were significantly different in LUSC and LUAD patients compared with the control group (all P < 0.05). The comparative analysis of clinical baseline data showed no obvious difference in age, gender, smoking, pack-years, tumor size, TNM stage, lymph node metastasis, distal metastasis, CEA level and comorbidities between LUSC and LUAD patients (all P > 0.05) (Table 2).
 |
Table 2 Comparative Analysis of Clinical Data Between NSCLC Patients and Healthy Subjects |
High Expression of lncRNA HEIH in Peripheral Blood of NSCLC Patients
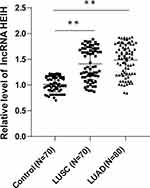
We detected the expression of lncRNA HEIH in peripheral blood of LUSC, LUAD patients and healthy subjects by qRT-PCR. LncRNA HEIH in peripheral blood of LUSC and LUAD patients was obviously higher than that of healthy controls (all P < 0.01), with no significant difference between LUSC and LUAD groups (P > 0.05) (Figure 1).
Correlation Analysis Between lncRNA HEIH Levels and Clinical Indexes of NSCLC Patients
To further study the relationship between lncRNA HEIH expression and clinical indicators of NSCLC patients, we assigned LUSC and LUAD patients into lncRNA HEIH low expression group and lncRNA HEIH high expression group according to lncRNA HEIH median level in LUSC and LUAD. In LUSC and LUAD patients, there was no evident difference in age, gender, smoking, and comorbidities between the lncRNA HEIH low and high expression groups, while the lncRNA HEIH high expression group had larger pack-years, tumor size, higher tumor stage and higher CEA level, and higher risk of lymph node metastasis and distal metastasis (all P < 0.05) (Table 3).
 |
Table 3 Correlation Analysis of lncRNA HEIH Expression in Peripheral Blood of NSCLC Patients and Clinical Indicators |
In addition, Logistic regression was conducted to analyze the factors influencing the expression of lncRNA HEIH in peripheral blood of NSCLC patients. With lncRNA HEIH expression as a dependent variable, the pack-years, tumor size, TNM stage, lymph node metastasis, distant metastasis, and CEA (P < 0.1) in Table 3 were incorporated into the binary Logistic regression equation as independent variables. The results showed that TNM stage, lymph node metastasis, distal metastasis, and CEA were independent risk factors affecting the expression of lncRNA HEIH in LUSC and LUAD (all P < 0.05) (Table 4).
 |
Table 4 Logistic Multifactor Regression Analysis of Influencing lncRNA HEIH Expression |
 |
Table 5 Meaning of Variable and Assignment Method |
 |
Table 6 Cox Multivariate Analysis of Independent Factors Influencing NSCLC Prognosis |
LncRNA HEIH Has High Diagnostic Value in NSCLC Patients
Tumor marker CEA has been reported as a biomarker for the auxiliary diagnosis and treatment effect of NSCLC.28–31 We evaluated the diagnostic efficacy of lncRNA HEIH and CEA in LUSC and LUAD patients through ROC curve analysis. The area under ROC curve of CEA in the diagnosis of LUSC patients was 0.706, the sensitivity was 31.43%, and the specificity was 100.00%. The area under ROC curve for lncRNA HEIH in the diagnosis of LUSC patients was 0.860, the sensitivity was 72.86%, and the specificity was 95.71% (Figure 2A). MedCalc-comparison of ROC curves showed that the area under ROC curve of lncRNA HEIH was significantly higher than that of CEA (P = 0.0031; 95% CI = 0.052~0.255), indicating that lncRNA HEIH had a higher diagnostic efficiency for LUSC than CEA. In addition, the area under ROC curve of lncRNA HEIH in the diagnosis of LUAD patients was 0.905, the sensitivity was 77.50%, and the specificity was 95.71%. The area under ROC curve of LUAD patients diagnosed by CEA was 0.763, the sensitivity was 50.00%, and the specificity was 90.00% (Figure 2B). MedCalc-comparison of ROC curves showed that the area under ROC curve of lncRNA HEIH was significantly higher than that of CEA (P = 0.0011; 95% CI = 0.057–0.228), indicating that lncRNA HEIH had a higher diagnostic efficacy than CEA for LUAD. The above data indicate that lncRNA HEIH has a high diagnostic efficacy in NSCLC patients.
High Expression of lncRNA HEIH Predicts Poor Prognosis of NSCLC
Furthermore, we analyzed the prognostic value of lncRNA HEIH in peripheral blood on NSCLC. According to the median level of lncRNA HEIH in LUSC and LUAD, patients with LUSC and LUAD were divided into lncRNA HEIH low expression group and lncRNA HEIH high expression group. Patients with NSCLC after the operation were followed up every 3 months for 60 months and the survival of the patients was recorded. The follow-up results showed that during the follow-up period, a total of 27 LUSC patients died at the end of the follow-up, including 18 cases in the high expression group and 9 cases in the low expression group. The cumulative survival rate in the lncRNA HEIH high expression group was evidently lower than that in the low expression group (P = 0.0252) (Figure 3A). A total of 34 LUAD patients died at the end of follow-up, including 24 cases in the high expression group and 10 cases in the low expression group. The cumulative survival rate in the lncRNA HEIH high expression group was notably lower than that of the low expression group (P = 0.0027) (Figure 3B). These results suggest that lncRNA HEIH overexpression predicts poor prognosis in patients with NSCLC.
LncRNA HEIH Expression Was an Independent Prognostic Risk Factor for NSCLC
To further evaluate the effect of lncRNA HEIH on the prognosis of NSCLC, we took the survival of patients as the dependent variable, and included age, smoking, pack-years, tumor diameter, TNM stage, lymph node metastasis, distal metastasis, CEA, comorbidities and LncRNA HEIH as independent variables into Cox multivariate regression analysis model according to the analysis in Table 3 and the possible risk factors affecting the prognosis of NSCLC as previously reported.32 The results showed that lncRNA HEIH was an independent prognostic risk factor for LUSC patients after adjustment for pack-years and CEA level (P=0.035, HR=9.752, 95% CI: 1.175–80.898); After adjusting for pack-years and CEA level, lncRNA HEIH was an independent prognostic risk factor for LUAD patients (P=0.034, HR=6.623, 95% CI: 1.156–37.950) (Tables 5 and 6).
Discussion
LncRNAs are important in gene regulation.33 LncRNA interacts with many transcription factors and affects lung cancer growth and spread.13,34 To date, many lncRNAs have been shown to be associated with the diagnosis and prognosis of NSCLC.35 It was reported that lncRNA DNS-AS1 up-regulated the protein level of anti-apoptotic factor Bcl-2 and promoted the growth, migration, and invasion of LUAD cells.36,37 Meanwhile, lncRNA XIST showed carcinogenic properties in NSCLC by regulating miR-449a and Bcl-2.38 Moreover, knockdown of lncRNA NEAT 1 attenuated the expression of hypoxia-inducible factor 1, thus affecting the tumor-like phenotype of pulmonary bronchial epithelial cells.39 HEIH is a lncRNA22 originally found in HBV-induced hepatocellular carcinoma, and is highly expressed in NSCLC tissues and cell lines, which can promote the proliferation and metastasis of NSCLC.24 This paper highlighted that the high expression of lncRNA HEIH in peripheral blood of NSCLC patients can assist in the diagnosis of NSCLC and predict poor prognosis.
A total of 220 subjects were included in this study, including 70 healthy subjects, 70 patients with LUSC, and 80 patients with LUAD. As a biomarker, tumor marker CEA can play a role as a predictor and prognostic factor in cancer patients.40,41 Intraoperative CEA monitoring can provide more valuable prognostic information for patients with LUAD, and patients with normal or elevated postoperative CEA level have worse overall survival than those with normal preoperative CEA level.42 Stage-specific embryonic antigen-4 was reported to be expressed in basal-like lung cancer and is associated with poor prognosis.32 Smoking cessation therapy can obviously improve lung function parameters and reduce serum CEA levels.43 However, CEA as a traditional tumor marker has limited sensitivity and specificity, and miRNAs have higher diagnostic and prognostic value.44 In the present study, serum CEA levels in LUSC and LUAD patients were higher than those in controls. Consistently, CEA has been identified as a biomarker to assist in the diagnosis of NSCLC.30,31 Taken together, high serum CEA level can be used for the preliminary diagnosis of NSCLC.
Next, it was found that LncRNA HEIH in peripheral blood of LUSC and LUAD patients was higher than that of healthy controls, while lncRNA HEIH expression between LUSC and LUAD groups had no significant difference. This result is consistent with previous reports that lncRNA HEIH is highly expressed in NSCLC tissues and cell lines.24 Briefly, lncRNA HEIH level can be used as a potential diagnostic indicator for NSCLC, but it is not yet clear to distinguish LUSC from LUAD. To further study the relationship between lncRNA HEIH expression and clinical indicators in NSCLC patients, LUSC and LUAD patients were divided into a low-expression group and a high-expression group, the lncRNA HEIH overexpression group had larger tumor size, higher tumor stage and higher CEA levels, and higher risk of lymph node metastasis and distal metastasis. Similarly, lncRNA-HEIH is highly expressed in melanoma tissues and cell lines, which is associated with late clinical stage and predicts poor prognosis in melanoma patients.25 LncRNA HEIH high expression in gastric cancer patients is closely related to medium-high differentiation, distant metastasis, lymph node metastasis, and deeper tumor invasion.45 Altogether, high expression of lncRNA HEIH is associated with poorer clinical indicators and higher cancer staging.
The tumor marker CEA has been well established as a biomarker for the diagnosis and treatment of NSCLC.28,29 We evaluated the diagnostic efficacy of lncRNA HEIH and CEA in patients with LSC and LUAD through ROC curve analysis, which manifested that lncRNA HEIH had a high diagnostic efficacy in patients with NSCLC. Our paper may identify a more effective biomarker for NSCLC diagnosis.
Further, we analyzed the prognostic value of lncRNA HEIH in NSCLC. We assigned LUSC and LUAD patients into a low-expression group and a high-expression group and then followed up the patients. As expected, the cumulative survival rate in patients with high expression of lncRNA HEIH was lower than that in the patients with low expression of lncRNA HEIH in LUSC and LUAD. In short, high expression of lncRNA HEIH predicts poor prognosis in patients with NSCLC. The expression of HEIH is up-regulated in ovarian cancer tissues and cell lines, and high expression of HEIH indicates a poor prognosis.46 Oesophageal squamous cell carcinoma patients with high lncRNA HEIH expression have poorer prognosis than those with low expression.47 These results are consistent with the trend of our results.
Conclusion
In conclusion, the high expression of lncRNA HEIH in peripheral blood is helpful to the diagnosis and prognosis prediction of NSCLC, and may provide a new reference for evaluation of NSCLC clinically. Nevertheless, due to the small number of cases and events included in this study, it is necessary to further expand the sample size to further clarify the diagnostic and prognostic ability of lncRNA HEIH. Moreover, the efficacy of predicting lung cancer only by detecting the expression of lncRNA HEIH in peripheral blood is limited, and more studies are still necessary to find suitable combined diagnostic markers. In addition, the role of lncRNA HEIH in the occurrence and development of NSCLC is still poorly understood, and further studies are needed. In future studies, we should carry out a larger multi-center study, expand the sample size and match the control to increase the credibility of the results. Meanwhile, we should further study the combined diagnostic and prognostic value of lncRNA HEIH and CEA as well as other biomarkers in NSCLC patients. More studies are required to explore the molecular regulatory mechanism of lncRNA HEIH in the occurrence and development of NSCLC.
Data Sharing Statement
All the data generated or analyzed during this study are included in this published article.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi:10.3322/caac.21492
2. Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi:10.1016/S0140-6736(16)30958-8
3. Ettinger DS, Wood DE, Aggarwal C, et al. NCCN guidelines insights: non-small cell lung cancer, version 1.2020. J Natl Compr Canc Netw. 2019;17:1464–1472. doi:10.6004/jnccn.2019.0059
4. Zhang J, Zhu N, Chen X. A novel long noncoding RNA LINC01133 is upregulated in lung squamous cell cancer and predicts survival. Tumour Biol. 2015;36:7465–7471. doi:10.1007/s13277-015-3460-9
5. Kim L, Kim KH, Yoon YH, et al. Clinicopathologic and molecular characteristics of lung adenocarcinoma arising in young patients. J Korean Med Sci. 2012;27:1027–1036. doi:10.3346/jkms.2012.27.9.1027
6. Zhao S, Guo X, Mizutani KI, et al. Overexpression of teashirt homolog 2 suppresses cell proliferation and predicts the favorable survival of lung adenocarcinoma. Int J Med Sci. 2021;18:1980–1989. doi:10.7150/ijms.52109
7. Saito S, Espinoza-Mercado F, Liu H, Sata N, Cui X, Soukiasian HJ. Current status of research and treatment for non-small cell lung cancer in never-smoking females. Cancer Biol Ther. 2017;18:359–368. doi:10.1080/15384047.2017.1323580
8. Yang Y, Wang M, Liu B. Exploring and comparing of the gene expression and methylation differences between lung adenocarcinoma and squamous cell carcinoma. J Cell Physiol. 2019;234:4454–4459. doi:10.1002/jcp.27240
9. Fu L, Wang H, Wei D, et al. The value of CEP55 gene as a diagnostic biomarker and independent prognostic factor in LUAD and LUSC. PLoS One. 2020;15:e0233283. doi:10.1371/journal.pone.0233283
10. Souza CP, Cinegaglia NC, Felix TF, et al. Deregulated microRNAs are associated with patient survival and predicted to target genes that modulate lung cancer signaling pathways. Cancers. 2020;12:2711. doi:10.3390/cancers12092711
11. Beylerli OA, Azizova ST, Konovalov NA, Akhmedov AD, Gareev IF, Belogurov AA. [Non-coding RNAs as therapeutic targets in spinal cord injury]. Zh Vopr Neirokhir Im N N Burdenko. 2020;84:104–110. Russian. doi:10.17116/neiro202084031104
12. Lorenzi L, Avila Cobos F, Decock A, et al. Long noncoding RNA expression profiling in cancer: challenges and opportunities. Genes Chromosomes Cancer. 2019;58:191–199. doi:10.1002/gcc.22709
13. Jiao PF, Tang PJ, Chu D, Li YM, Xu WH, Ren GF. Long non-coding RNA THOR depletion inhibits human non-small cell lung cancer cell growth. Front Oncol. 2021;11:756148. doi:10.3389/fonc.2021.756148
14. Wang T, Yang C, Li B, et al. Identification of lncRNA-miRNA-mRNA networks linked to non-small lung cancer resistance to inhibitors of epidermal growth factor receptor. Front Genet. 2021;12:758591. doi:10.3389/fgene.2021.758591
15. Song H, Li H, Ding X, et al. Long noncoding RNA FEZF1AS1 facilitates nonsmall cell lung cancer progression via the ITGA11/miR516b5p axis. Int J Oncol. 2020;57:1333–1347. doi:10.3892/ijo.2020.5142
16. Yang J, Qiu Q, Qian X, et al. Long noncoding RNA LCAT1 functions as a ceRNA to regulate RAC1 function by sponging miR-4715-5p in lung cancer. Mol Cancer. 2019;18:171. doi:10.1186/s12943-019-1107-y
17. Shi X, Liu Z, Liu Z, et al. Long noncoding RNA PCAT6 functions as an oncogene by binding to EZH2 and suppressing LATS2 in non-small-cell lung cancer. EBioMedicine. 2018;37:177–187. doi:10.1016/j.ebiom.2018.10.004
18. Reis EM, Verjovski-Almeida S. Perspectives of long non-coding RNAs in cancer diagnostics. Front Genet. 2012;3:32. doi:10.3389/fgene.2012.00032
19. Luo J, Li Q, Pan J, Li L, Fang L, Zhang Y. Expression level of long noncoding RNA H19 in plasma of patients with nonsmall cell lung cancer and its clinical significance. J Cancer Res Ther. 2018;14:860–863. doi:10.4103/jcrt.JCRT_733_17
20. Tan J, Yang L. Long noncoding RNA VPS9D1-AS1 overexpression predicts a poor prognosis in non-small cell lung cancer. Biomed Pharmacother. 2018;106:1600–1606. doi:10.1016/j.biopha.2018.07.113
21. Xie Y, Zhang Y, Du L, et al. Circulating long noncoding RNA act as potential novel biomarkers for diagnosis and prognosis of non-small cell lung cancer. Mol Oncol. 2018;12:648–658. doi:10.1002/1878-0261.12188
22. Yang F, Zhang L, Huo XS, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi:10.1002/hep.24563
23. Cui C, Zhai D, Cai L, Duan Q, Xie L, Yu J. Long noncoding RNA HEIH promotes colorectal cancer tumorigenesis via counteracting miR-939Mediated transcriptional repression of Bcl-xL. Cancer Res Treat. 2018;50:992–1008. doi:10.4143/crt.2017.226
24. Jia K, Chen F, Xu L. Long noncoding RNA HEIH promotes the proliferation and metastasis of non-small cell lung cancer. J Cell Biochem. 2019;120:3529–3538. doi:10.1002/jcb.27629
25. Zhao H, Xing G, Wang Y, Luo Z, Liu G, Meng H. Long noncoding RNA HEIH promotes melanoma cell proliferation, migration and invasion via inhibition of miR-200b/a/429. Biosci Rep. 2017;37. doi:10.1042/BSR20170682
26. Beasley MB, Brambilla E, Travis WD. The 2004 World Health Organization classification of lung tumors. Semin Roentgenol. 2005;40:90–97. doi:10.1053/j.ro.2005.01.001
27. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi:10.1038/nprot.2008.73
28. Clevers MR, Kastelijn EA, Peters BJM, Kelder H, Schramel F. Evaluation of serum biomarker CEA and Ca-125 as immunotherapy response predictors in metastatic non-small cell lung cancer. Anticancer Res. 2021;41:869–876. doi:10.21873/anticanres.14839
29. Dal Bello MG, Filiberti RA, Alama A, et al. The role of CEA, CYFRA21-1 and NSE in monitoring tumor response to Nivolumab in advanced non-small cell lung cancer (NSCLC) patients. J Transl Med. 2019;17:74. doi:10.1186/s12967-019-1828-0
30. Li C, Lv Y, Shao C, et al. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J Cell Physiol. 2019;234:20721–20727. doi:10.1002/jcp.28678
31. Li Q, Sang S. Diagnostic value and clinical significance of combined detection of serum markers CYFRA21-1, SCC Ag, NSE, CEA and ProGRP in non-small cell lung carcinoma. Clin Lab. 2020;66. doi:10.7754/Clin.Lab.2020.191243
32. Zamarron E, Prats E, Tejero E, et al. Static lung hyperinflation is an independent risk factor for lung cancer in patients with chronic obstructive pulmonary disease. Lung Cancer. 2019;128:40–46. doi:10.1016/j.lungcan.2018.12.012
33. Wang M, Dai M, Wang D, et al. The long noncoding RNA AATBC promotes breast cancer migration and invasion by interacting with YBX1 and activating the YAP1/Hippo signaling pathway. Cancer Lett. 2021;512:60–72.
34. He Y, Jiang X, Duan L, et al. LncRNA PKMYT1AR promotes cancer stem cell maintenance in non-small cell lung cancer via activating Wnt signaling pathway. Mol Cancer. 2021;20:156. doi:10.1186/s12943-021-01469-6
35. Zhang C, Gong C, Li J, Tang J. Downregulation of long non-coding RNA LINC-PINT serves as a diagnostic and prognostic biomarker in patients with non-small cell lung cancer. Oncol Lett. 2021;21:210. doi:10.3892/ol.2021.12471
36. Liu Y, Liang L, Ji L, et al. Potentiated lung adenocarcinoma (LUAD) cell growth, migration and invasion by lncRNA DARS-AS1 via miR-188-5p/ KLF12 axis. Aging. 2021;13:23376–23392. doi:10.18632/aging.203632
37. Sun Y, Hu B, Wang Q, et al. Long non-coding RNA HOTTIP promotes BCL-2 expression and induces chemoresistance in small cell lung cancer by sponging miR-216a. Cell Death Dis. 2018;9:85. doi:10.1038/s41419-017-0113-5
38. Zhang YL, Li XB, Hou YX, Fang NZ, You JC, Zhou QH. The lncRNA XIST exhibits oncogenic properties via regulation of miR-449a and Bcl-2 in human non-small cell lung cancer. Acta Pharmacol Sin. 2017;38:371–381. doi:10.1038/aps.2016.133
39. Jiang P, Hao S, Xie L, et al. LncRNA NEAT1 contributes to the acquisition of a tumor like-phenotype induced by PM 2.5 in lung bronchial epithelial cells via HIF-1alpha activation. Environ Sci Pollut Res Int. 2021;28:43382–43393. doi:10.1007/s11356-021-13735-7
40. Iacuzzo C, Germani P, Troian M, et al. Serum carcinoembryonic antigen pre-operative level in colorectal cancer: revisiting risk stratification. ANZ J Surg. 2021;91:E367–E374.
41. Zhang M, Huang C, Zhou H, et al. Circulating tumor DNA predicts the outcome of chemotherapy in patients with lung cancer. Thorac Cancer. 2021;13:95–106.
42. Jiao Z, Cao S, Li J, et al. Clinical associations of preoperative and postoperative serum CEA and lung cancer outcome. Front Mol Biosci. 2021;8:686313. doi:10.3389/fmolb.2021.686313
43. Pezzuto A, Spoto C, Vincenzi B, Tonini G. Short-term effectiveness of smoking-cessation treatment on respiratory function and CEA level. J Comp Eff Res. 2013;2:335–343. doi:10.2217/cer.13.25
44. Yaghoubi N, Avval FZ, Khazaei M, Sahebkar A, Aghaee-Bakhtiari SH. High diagnostic and prognostic value of miRNAs compared with the carcinoembryonic antigen as a traditional tumor marker. Anticancer Agents Med Chem. 2021. doi:10.2174/1871520621666210608094908
45. Chen X, Sun X, Li X, Xu L, Yu W. LncRNA-HEIH is a novel diagnostic and predictive biomarker in gastric cancer. Genet Test Mol Biomarkers. 2021;25:284–292. doi:10.1089/gtmb.2020.0270
46. Si L, Chen J, Yang S, et al. lncRNA HEIH accelerates cell proliferation and inhibits cell senescence by targeting miR-3619-5p/CTTNBP2 axis in ovarian cancer. Menopause. 2020;27:1302–1314. doi:10.1097/GME.0000000000001655
47. Ding X, Qi C, Min J, Xu Z, Huang K, Tang H. Long non-coding RNA HEIH suppresses the expression of TP53 through enhancer of zeste homolog 2 in oesophageal squamous cell carcinoma. J Cell Mol Med. 2020;24:10551–10559. doi:10.1111/jcmm.15673
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.