Back to Journals » OncoTargets and Therapy » Volume 14
High Expression of LINC01268 is Positively Associated with Hepatocellular Carcinoma Progression via Regulating MAP3K7
Authors Jin X, Fu WX, Li D, Wang N, Chen J, Zeng Z, Guo J, Liu H, Zhong X, Peng H, Yu X, Sun J, Zhang X, Wang X, Xu B, Lin Y, Liu J , Kutter C, Li Y
Received 4 December 2020
Accepted for publication 22 February 2021
Published 8 March 2021 Volume 2021:14 Pages 1753—1769
DOI https://doi.org/10.2147/OTT.S295814
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Xiuli Jin,1 Weixin Fu,2 Dan Li,1 Ningning Wang,1 Jiayu Chen,1 Zilu Zeng,1 Jiaqi Guo,1 Hao Liu,3 Xinping Zhong,3 Hu Peng,4 Xin Yu,5 Jing Sun,1 Xinhe Zhang,1 Xue Wang,1 Beibei Xu,1 Yingbo Lin,6 Jianping Liu,4 Claudia Kutter,7 Yiling Li1
1Department of Gastroenterology, First Affiliated Hospital of China Medical University, Shenyang, 110001, People’s Republic of China; 2Science Experiment Center of China Medical University, Shenyang, 110122, People’s Republic of China; 3Department of Hepatobiliary Surgery, First Affiliated Hospital of China Medical University, Shenyang, 110001, People’s Republic of China; 4Emergency Department, Shanghai Tenth People’s Hospital, School of Medicine, Tongji University, Shanghai, 200072, People’s Republic of China; 5Department of Human Anatomy, School of Basic Medicine, Dali University, Dali, Yunnan, 671003, People’s Republic of China; 6Department of Oncology-Pathology, Karolinska Institute, Stockholm, 17177, Sweden; 7Department of Microbiology, Tumor and Cell Biology, Science for Life Laboratory, Karolinska Institute, Stockholm, 17177, Sweden
Correspondence: Claudia Kutter
Department of Microbiology, Tumor and Cell Biology, Science for Life Laboratory, Karolinska Institute, Stockholm, 17177, Sweden
Email [email protected]
Yiling Li
Department of Gastroenterology, First Affiliated Hospital of China Medical University, Shenyang, 110001, People’s Republic of China
Email [email protected]
Objective: As one of the most common neoplastic diseases, hepatocellular carcinoma (HCC) has a high morbidity and mortality, which seriously threatens human health and places a heavy burden on society and medical care. At present, effective early diagnosis, prognosis and treatment of HCC are limited. Altered gene expression patterns of lncRNA are associated with the occurrence, development and prognosis of various malignancies, including HCC. The aim of this study was to investigate the correlation between the expression of LINC01268 and HCC, and to elucidate the potential underlying molecular mechanism.
Methods: Expression level and localization of LINC01268 in human liver cancer cells and HCC tissues were investigated using RT-qPCR and fluorescent in situ hybridization (FISH), respectively. Correlation of expression levels of LINC01268 and MAP3K7 with differentiation and poor overall patient survival of HCC were analyzed using in house collected and publicly available HCC tissue data. RT-qPCR and Western blot were applied to inspect the effects of depletion and overexpression of LINC01268 on MAP3K7 expression. HCC cell proliferation and apoptosis were also investigated by simultaneous overexpression of LINC01268 and knockdown of MAP3K7, in order to delineate that MAP3K7 is a downstream effector of LINC01268.
Results: In this study, we identified that LINC01268 was highly expressed in HCC cell lines and tissues. High LINC01268 expression level was associated with lower HCC nodule number, moderate/poor differentiation and poor overall survival. Knockdown of LINC01268 inhibited the proliferation of HCC cells, which was enhanced by overexpression of LINC01268. Co-expression analysis implied an interaction between LINC01268 and MAP3K7. Similar to LINC01268, MAP3K7 was highly expressed in HCC cells, and positively correlated with moderate/poor differentiation as well as poor prognosis. Knockdown of LINC01268 in HCC cell lines led to reduction of MAP3K7 at both mRNA and protein levels. Phenotypic effects due to LINC01268 overexpression in HCC cells were reversed by knockdown of MAP3K7.
Conclusion: Taken together, the abnormal high expression of LINC01268 is associated with HCC progression via regulating MAP3K7, suggesting LINC01268 as a novel marker for HCC prognosis and potentially a new therapeutic target.
Keywords: LINC01268, MAP3K7, hepatocellular carcinoma, proliferation, prognosis
Introduction
With 600,000 death per year, hepatocellular carcinoma (HCC) is the fourth most common cause of cancer-related deaths worldwide.1 China has the highest number of incidences and accounts for about half of the new cases of HCC every year. Intensive work has been conducted to identify HCC risk factors (eg, Hepatitis virus infections, non-alcoholic fatty liver disease and obesity, smoking and alcohol as well as the genetic background) and to implement preventive measurements.2 At present, despite considerable progress in HCC prevention, diagnosis and intervention, only 20% of the patients survive more than one year after diagnosis.3 A number of serological biomarkers have been widely used in diagnosis of HCC,4–6 such as alpha-fetoprotein (AFP), alpha-L-fucosidase (AFU), gamma-glutamyl transferase (GGT), des-gamma-carboxy prothrombin (DCP), glypican-3 (GPC3) and golgi glycoprotein 73 (GP73). However, due to limited specificity and sensitivity, new biomarkers are required, which in combination with the current biomarkers can better refine HCC diagnosis and prognosis,7 develop new interventions and treatment strategies.
Large-scale gene expression analyses have described pervasive gene transcriptions, which are commonly linked to deregulation of noncoding and protein-coding genes in biopsies of cancer patients and derivative cell lines.8 Noncoding gene expression accounts for more than 98% of all gene products in the human genome.9 Long noncoding RNAs (lncRNAs) are a group of noncoding genes and annotated intensively over the past years. Despite tremendous efforts to catalogue lncRNA genes, it remains challenging to assign functionality since lncRNAs are often expressed in a species-, spatiotemporal-, cellular- and tissue-specific manner.10 However, detailed studies of selected cases revealed that lncRNAs could interact with DNA, RNA and proteins and thereby regulate various molecular processes ranging from gene expression to protein translation.11 Given these versatile modes of actions, lncRNAs can thereby influence crucial cellular responses that define eg cell differentiation, organ formation and pathological changes among others. Furthermore, due to the restricted expression patterns, lncRNAs can be used to pinpoint varying degrees of tumor malignancies.12 Hence, abnormal gene expression patterns of lncRNAs have received extensive attentions in liver cancer research in recent years.13,14 For example, lncRNA-ATB,11,15 lncRNA HULC,16–19 lncRNA HOTTIP and lncRNA HOXAB,20 lncRNA HOTAIR,21 lncRNA CUDR,22 lncTCF7,23 lncRNA NEAT1,24–26 lncRNA MT1DP,27 lncCAMTA128 and lncDILC29 have been demonstrated to be involved in the occurrence, development and prognosis of HCC.30,31
Previous reports indicated that high expression of LINC01268 is related to suicide by violent means,32,33 glioma34 and acute myeloid leukemia.35 In this study, we identified and further investigated the regulatory role of LINC01268 (also known as LOC285758, ROCKI or MROCKI) in HCC liver biopsies and cell lines. Our data indicated that LINC01268 regulate gene expression of mitogen-activated protein kinase kinase kinase 7 (MAP3K7, MEKK7, TAK1). Both LINC01268 and MAP3K7 are highly abundant in HCC liver biopsies and cell lines, other than in peritumoral tissues or normal liver cells. Of clinical relevance, we showed that high gene expression levels of LINC01268 and MAP3K7 are positively associated with medium to poor HCC differentiation and poor overall patient survival. On the cellular level, we showed that knockdown of LINC01268 reduced MAP3K7 abundance on the mRNA and protein levels. Phenotypically, we observed that reduction of LINC01268 expression repressed proliferation of HCC cells, which could be facilitated via the NF-κB36 and MAPK37,38 pathways. On the other hand, enhanced proliferation of HCC cells due to LINC01268 overexpression were reversed by knockdown of MAP3K7. In conclusion, LINC01268 could be a novel marker for the prognosis of HCC and a potential therapeutic target.
Materials and Methods
Ethical Statement
Informed consents were obtained from all patients and all experiments with tissue samples were carried out in accordance with the Declaration of Helsinki and the guidelines of the Ethics Committee of the First Affiliated Hospital of China Medical University (License number 2019–172-4).
Collection of Tissue Samples from HCC Patients
Forty-six pairs of HCC and adjacent para-cancerous tissues were −80°C frozen after collection from patients with their consent at the Department of Hepatobiliary Surgery, the First Affiliated Hospital of China Medical University between April 2018 and May 2019, among which 18 pairs were used for analyses of both LINC01268 and MAP3K7 (Supplementary Table 1, not highlighted), 14 pairs only for LINC01268 analysis (Supplementary Table 1, highlighted in blue) and the other 14 pairs only for MAP3K7 analysis (Supplementary Table 1, highlighted in green). These patients did not receive any treatments before surgery. HCC patient diagnosis was confirmed by histopathology. Detailed patient descriptions and their pathological assessments, including gender, age, tumor size, HCC nodule number, metastasis, pathological type, tumor differentiation, HBV infection status, cirrhosis and concentration of Alpha Fetoprotein (AFP, ng/mL) are listed in Supplementary Table 1.
Cell Culture
Human fetal hepatocyte cell line LO2, liver hepatocellular carcinoma cell lines HepG2, Huh7 and Hep3B, lung cancer cell line A549 and breast cancer cell line T47D were purchased from commercial source (Cell Bank of Chinese Academy of Sciences) and tested to be mycoplasma negative (LookOut® Mycoplasma PCR Detection Kit, Cat. No.: MP0035, Sigma-Aldrich) prior to this study. After identity confirmation using GoldeneyeTM 20A STR complex Amplification Kit, the cells were grown to approximate 80% confluence in Dulbecco’s modified Eagle’s medium with high glucose (HyClone) supplemented with 10% fetal bovine serum (FBS) (Gibco), 100 U penicillin and 100 μg/mL streptomycin (Gibco) at 37°C and 5% CO2.
Fluorescent in situ Hybridization (FISH)
Three paired HCC and corresponding adjacent para-cancerous tissues were randomly selected as representatives, embedded in paraffin and cut into 8 μm tissue slices. LO2, HepG2 and Huh7 cells grown on coverslips were fixed by 4% paraformaldehyde (20 min, room temperature) and washed with 1×PBS (3×5 min, room temperature). After digestion with proteinase K (20 μg/mL) for 8 min, washing with deionized water and 1×PBS (3×5 min, room temperature), tissue and cell samples were incubated with pre-hybridization solution (Servicebio, Wuhan, G3016-3) in a 37°C incubator for 1 h. Subsequently, after removing the pre-hybridization solution, the hybridization solution (Servicebio, Wuhan, containing 6 ng/μL of the LINC01268 probe, Table 1) was added onto the slides and incubated at 37°C overnight. The samples were then washed successively with 2×SSC for 10 min, 1×SSC for 2×5 min and 0.5×SSC for 10 min at 37°C. Next, the samples were blocked at room temperature for 30 min, followed by incubation with anti-DIG-HRP antibody (1:300 dilution, Jackson ImmunoResearch) for 50 min and washed with 1×PBS for 3×5 min at 37°C. Afterwards, the FITC-TSA reagent (1:3000 dilution, Servicebio) was applied for incubation at room temperature in darkness for 5 min, followed by washing with 1×PBS for 3×5 min, incubation with aqueous fluoroshield mounting medium with DAPI (2 μg/mL, Abcam, ab104139) for nuclei staining in darkness for 8 min.
 |
Table 1 Oligo Sequences Used in This Study |
Images were acquired using a Nikon Eclipse Ci-L upright fluorescence microscope (Nikon Instruments) and imaging system NIKON DS-U3. Nuclei and LINC01268 were visualized in blue (excitation 330–380 nm, emission 420 nm) and green (excitation 465–495 nm, emission 515–555 nm), respectively.
RNA Extraction and RT-qPCR
Total RNA from frozen tissues and cell lines were extracted using the RNAiso Plus RNA isolation kit (Takara) according to the manufacturer’s instruction with DNase treatment. Five hundred nanogram of total RNA was reverse transcribed using oligo (dT) primer (Primer Script RT, Takara) for subsequent qPCR (SYBR Primer Ex Taq II, Clontech) with gene-specific primers (Table 1) on LightCycler480 (Applied Biosystems). The relative expression was calculated using the ΔΔCt method. β-ACTIN served as an internal control. For technical reproducibility, the experiment was independently repeated in triplicate.
Cellular Transfection
HCC cells (HepG2, Hep3B and Huh7) were seeded into 96-well plates (5000 cells/well) or 6-well plates (3.5 × 105 cells/well) upon transfection with Lipofectamine 3000 (Thermofisher Scientific) (0.15 μL/well for 96-well plate or 7.5 μL/well for 6-well plates) and 100 nM siRNAs (Guangzhou Ruibo Biotechnology Co., Table 1). As shown in Results section that LINC01268 is located in both cytoplasm and nuclei, a mixture of siLINC01268 Smart silencers targeting LINC01268 comprising of three siRNA sequences (silencer 1–3, double-stranded RNA complex, with target sequences shown in Table 1) and three antisense oligonucleotides (silencer 4–6, single-stranded DNA, with target sequences shown in Table 1) was applied to down-regulate the expression of LINC01268. Non-targeting siCtrl served as the control for data normalization and comparison. The knockdown efficiency was determined two days post transfection. Each treatment was performed in three independent biological replicates.
Apoptosis Assay
HepG2 and Huh7 were transfected in 6-well plates at a density of 1×105 cells/well. Two days post transfection, the cells were harvested and the apoptotic cells were analyzed using Annexin V- AbFlourTM 488 Apoptosis Detection Kit (Abbkine) according to the manufacturer’s instructions at LSRFortessa™ X-20 Cell Analyzer (Becton Dickinson, USA) with software CELLQuest Version 5.1.
Colorimetric Cell Proliferation Assay
HepG2 and Huh7 cells were plated into 96-well plates at a density of 5000 cells/well and transfected with siLINC01268 and siCtrl, respectively, as described above. Cell proliferation was measured on day 0, 1, 2, 3 and 4 using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS) (Promega) according to the manufacturer’s instructions. The absorbance at 490 nm was recorded using a 96-well plate reader (Infinite F200 PRO, Tecan) and software i-control (V1.5).
EdU Cell Proliferation Assay
Cell proliferation was analyzed by EdU (5-ethynyl-20- deoxyuridine) assay using a Cell-Light EdU DNA Cell Proliferation Kit (RiboBio) as previously described.39 HepG2, Huh7 and Hep2B cells were plated into 96-well plates at a density of 5000 cells/well and transfected with siLINC01268, siMAP3K7 or siCtrl, as described above. After incubation for 48 h, 50 mM EdU was added and incubated at 37°C for 2 h before fixation with 4% formaldehyde for 30 min and subsequent incubation with 2 mg/mL glycine for 5 min. After washing with 1×PBS for 5 times, the cells were stained with 100 μL/well of 1× Apollo reaction cocktail for 30 min to detect the proliferating cells. Finally, Hoechst 33,342 (5 μg/mL) was added to stain the nuclei. The images were acquired using the Olympus microscope BX63 (Olympus) and software cellSens (Version 3.1). Cell proliferation was analyzed according to the manufacturer’s instructions and quantified by ImageJ (Version 1.0.3).
Generation of HepG2 and Huh7 Cell Lines Stably Over-Expressing LINC01268
Full length cDNA of LINC01268 was amplified from Hep3B cells and ligated to over expression vector pEX-3(pGCMV/MCS/Neo) (Genepharma, Suzhou, China) after enzymatic digestion and gel purification. HepG2 and Huh7 cells (4×105/well) were transfected with either empty vector (as negative control) or LINC01268 overexpression plasmid in 6-well plates (3.75 μL Lipofectamine 3000 and 2.5 μg plasmid DNA per well), respectively. Single positive colonies were picked by G418 selection (200–400 μg/mL) and expanded to obtain the stable LINC01268-overexpressing HepG2 and Huh7 cells, which were subjected to further analyses, such as MAP3K7 knockdown, proliferation and apoptosis determination as described above.
Survival and Gene Co-Expression Analysis
We used the interactive web tool GEPIA40 (GEPIA2 2019 Release, http://gepia.cancer-pku.cn/about.html) that visualizes pre-computed data from The Cancer Genome Atlas (TCGA, https://www.cancer.gov/tcga)41 and the Genotype-Tissue Expression (GTEx, https://www.gtexportal.org)42 projects. Data used for the tissue-type specific identification of LINC01268 were obtained from the Genotype-Tissue Expression (GTEx) Portal on 2020–09-24. Survival plots of LINC01268 and MAPK3K7 were obtained by analyzing the liver hepatocellular carcinoma (LIHC) dataset using defaulted parameters. Similarly, we searched for co-expression of genes when querying for LINC01268 in TCGA LIHC tumor versus LIHC normal samples (Supplementary Table 2). Furthermore, GEPIA was also used to correlate gene expression levels of LINC01268 with MAP3K7.
Protein Immunoblotting
Total cellular proteins were extracted from the cells using RIPA Lysis Buffer and quantified using a BCA Protein Assay Kit (Beyotime). Protein samples (15 μg per lane) were separated on a 10% SDS-PAGE gel and transferred onto a polyvinylidene fluoride membrane (Millipore). After blocking, the membranes were incubated with anti-TAK1 (1:1000 dilution, ab109526, Abcam) and β-actin antibody (1:3000 dilution, abs132001, Absin) at 4°C overnight. After washing, the membranes were then incubated with secondary antibody (1:5000 dilution, HRP-conjugated Affinipure Goat Anti-Rabbit IgG (H+L), Proteintech) at room temperature for 1 hour. The blots were developed using a chemiluminescence system (Tanon‐4200, Tanon Science & Technology, Shanghai, China) and software GIS1D (Gel Image System Ver 4.00).
Statistical Analysis
The measurement data were expressed by mean ± SD. Statistical analysis was performed using SPSS 22.0 software. Student’s t-test was performed for p-value calculation to determine the significance of differential gene expression of LINC01268 and MAP3K7 between HCC samples and their corresponding adjacent para-cancerous tissues. A value of p<0.05 (marked with *) was considered significant, while p<0.01 (marked with **) was considered very significant and p<0.001 (marked with ***) was considered extremely significant.
Results
LINC01268 is Deregulated in HCC Patients
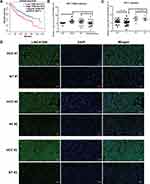
Cancer and cell-type-specific expression patterns of lncRNAs have been reported, which exemplified oncogenic and tumor suppressing functions of some lncRNAs.10–12,20,43 When we analyzed the bulk RNA-seq data (not shown here) from human liver cancer cell lines HepG2 and Huh7, together with some other human cancer cell lines and non-cancer cell lines, a number of polyA-containing LincRNAs were revealed to be specifically deregulated in these two human liver cancer cell lines. We choose LINC01268 for further functional studies after ranking the expression counts and RT-qPCR validation in three HCC cell lines (HepG2, Huh7 and Hep3B) and three non-HCC cell lines (LO2, A549 and T47D) used as controls (Figure 1A).
The primate-specific noncoding RNA LINC0126844 is more abundant in normal liver (Figure 1B, red arrow) as well as in lung, small intestine and colon tissue, compared to other profiled organs or tissues, suggesting organ-specific roles. However, the median expression level in liver is below five transcripts per million reads, with no obvious differences between genders. Remarkably, LINC01268 has been previously described to be upregulated in gliomas when compared to normal tissue34 and got assigned prognostic value for acute myeloid leukemia.35 We, therefore, sought to gain more knowledge of the role of LINC01268 in liver cancer. For that, we explored the correlation of LINC01268 expression level with the survival of liver cancer patients by examining the overall survival of a cohort of 362 HCC patients (LIHC) from The Cancer Genome Atlas (TCGA) database. The overall survival analysis showed that low gene expression of LINC01268 (181 cases) resulted in a significantly better patient survival outcome whereas high gene expression of LINC01268 (181 cases) is linked to poorer patient survival (p<0.01) (Figure 2A). The association between gene expression level and overall survival is consistent over shorter (0–60 months) and longer (60–120 months) time period after diagnosis, proposing that differential gene expression levels of LINC01268 has prognostic value over time. The decrease in patient survival upon higher gene expression of LINC01268 suggested that LINC01268 is linked to more malignant stages of HCC. Thus, we investigated LINC01268 expression levels in different liver cancer stages. The histological tumor grade was determined in 32 paired liver cancer biopsies and distinguished tumors that have some remaining phenotypical similarity to hepatocytes (“well differentiated”) (n=8) from entirely abnormal phenotypes that do not resemble any known cell type (“moderately or poorly differentiated”) (n=24). Our RT-qPCR results showed that LINC01268 expression was significantly lower (5.5 fold) in well-differentiated (2.95 fold of the corresponding surrounding non-tumor counterparts) when compared to moderately or poorly differentiated tumors (0.54 fold of the corresponding surrounding non-tumor counterparts) (Student’s t-test, p=0.042) (Figure 2B). This suggests that the LINC01268 expression can be used to discern the grade of tumor differentiation. Furthermore, the RT-qPCR analysis revealed that LINC01268 expression was significantly elevated in tumors with one nodule (n=29) compared to tumors with two or more nodules (n=3) (Student’s t-test, p=0.026) (Figure 2C). By inspecting additional physiological and clinical information from the HCC patients, we found that LINC01268 expression levels did not significantly correlate with age, gender, HBV infection status, serological Alpha Fetoprotein (AFP) levels, tumor size or occurrence of cirrhosis (Table 2).
 |
Table 2 Relationship Between LINC01268 Expression and Clinical Characteristics of HCC |
Overall, our results highlighted that LINC01268 expression level, in combination with established diagnostic parameters and tools, can be indicative for liver cancer malignancy.
LINC01268 Shows Distinct Spatial Gene Expression Pattern in HCC Cells
To further explore the relevance of LINC01268 as an indicator for HCC, we investigated the expression pattern and abundance of LINC01268 across three randomly selected paired HCC (moderate/poor differentiation) and matching adjacent para-cancerous tissues (Supplementary Table 1, highlighted in red) by fluorescence in situ hybridization (FISH). The FISH probe against LINC01268 (Table 1) resulted in green fluorescent signals (FITC), indicating that LINC01268 expression is detectable in liver tissues (Figure 2D). We noticed that LINC01268 expression is barely present in para-cancerous but highly abundant in moderately or poorly differentiated HCC tissues, which is consistent with our RT-qPCR data (Figure 2B). The expression of LINC01268 was uniformly distributed across the HCC tissue section. Similar to previous observations,44 we detected LINC01268 in the nucleus. In addition, cytoplasmic LINC01268 was also detected in our assay.
Our results show that LINC01268 can be used to discern healthy and cancerous liver cells. Its abundance in the nucleus and cytoplasm suggests a transcriptional and posttranscriptional role of LINC01268 in liver cancer.
Gene Silencing of LINC01268 in HCC Cells Inhibits Cell Proliferation and Promotes Cell Apoptosis
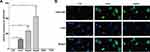
Given the higher prevalence of LINC01268 in liver cancer tissues (Figure 2), we next measured gene expression of LINC01268 in various cancer cell lines. In alignment with our observations from HCC tissues (Figure 2B), we determined by RT-qPCR that LINC01268 expression is significantly elevated in all three human HCC cell lines used in this study (Huh7, HepG2 and Hep3B) (Figure 3A). In contrast, LINC01268 expression was barely measurable in LO2, which resembles a more normal hepatocyte phenotype (Figure 3A). This result supported our observation that LINC01268 expression increased with tumor grade in patients (Figure 2B). Despite high gene expression in all HCC cells tested, we found varying degrees of LINC01268 abundance, for instance, Hep3B expressed LINC01268 2.7- and 1.9-fold more than Huh7 and HepG2, respectively (Figure 3A). LINC01268 expression was not detectable in lung (A549) or breast (T47D) cancer cell lines (Figure 3A), suggesting that the gene expression level of LINC01268 is cancer type specific. We next investigated the subcellular distribution of LINC01268 in LO2, HepG2 and Huh7 cells by FISH. In alignment to our results obtained from patient biopsies (Figure 2D), we found that LINC01268 is less abundant in non-malignant LO2 cells that resemble a more normal phenotype, but much higher in the HCC cell lines HepG2 and Huh7 (Figure 3B). Similar to the patient tissues, we found that LINC01268 RNA is present in both the nucleus and the cytoplasm. These results showed that the molecular phenotype observed in vivo in clinical tissues was resembled in vitro in HCC cell lines.
We further investigated the potential regulatory roles of altered LINC01268 expression levels. Two days post transfection in three HCC cell lines (Huh7, HepG2 and Hep3B) with silencer siLINC01268, we achieved a significant reduction of LINC01268 transcript levels when normalized to the siRNA control (siCtrl) (Figure 4A, 75% in Huh7, 70% in HepG2 and 53% in Hep3B, Student’s t-test, p<0.05). Remarkably, knockdown of LINC01268 expression resulted in a notable reduction of liver cancer cell proliferation as measured by EdU proliferation assay (Figure 4B and C, 62% in Huh7, 84% in HepG2 and 44% in Hep3B). Further detailed analysis in Huh7 and HepG2 confirmed a significant inhibition of cell proliferation within the first two days after knockdown (Figure 4D-E, Student’s t-test, p<0.05). Cell proliferation remained inhibited up to four days, although only significant in Huh7 but not in HepG2 for day 3 and 4 (Figure 4D-E). This could be explained by technical limitations (eg, relatively lower knockdown efficiency in HepG2 cells, Figure 4A) or undetermined cell line-dependent responses. Lastly, we measured the effects of the LINC01268 knockdown on apoptosis by FACS. We detected a significant increase of cell apoptosis in Huh7 and HepG2 upon LINC01268 knockdown (Figure 4F). In conclusion, the reduction of LINC01268 expression suppressed cell proliferation in vitro in HCC cell lines.
LINC01268 and MAP3K7 are Co-Expressed in HCC Liver Tissues and Cell Lines
A previous report highlighted the cis-regulatory role of LINC01268 in THP1-derived macrophages upon infection, whereby LINC01268 regulates the neighboring protein-coding gene myristoylated alanine-rich protein kinase C (MARCKS) and subsequently controls inflammatory cytokines and chemokine production.44 In order to identify regulatory interactions in liver cancer beyond this cis-acting function, we searched for co-expression patterns of LINC01268 with other genes in the GEPIA database40 and found that gene expression of LINC01268 correlated well with 60 genes (Supplementary Table 2, Pearson correlation coefficient, R ≥ 0.5). We found that the majority (55%, 33/60) of co-expressed genes were other annotated lncRNA and 45% (27/60) represented protein-coding genes, where we identified MAP3K7. Interestingly, previous reports described the involvement of MAP3K7 in HCC initiation and progression through various signaling pathways.45–49
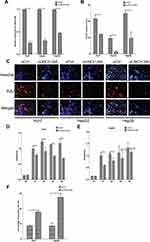
We found a strong correlation when inspecting expression levels of LINC01268 and MAP3K7 in the liver cancer patients (Figure 5A). Indeed, we noted that high expression of MAP3K7 was significantly associated with poor overall survival in HCC patients (p=0.02) (Figure 5B). Unlike LINC01268 (Figure 2A), high gene expression levels of MAP3K7 are only associated with poor survival within the initial time period (0–50 months) after diagnosis, suggesting that MAP3K7 is more likely to contribute to the early progression of HCC. Our RT-qPCR analysis revealed that MAP3K7 expression was much lower (6.9-fold) in well-differentiated cancer tissues (n=6) when compared to moderately or poorly differentiated cancer tissues (n=26) (Student’s t-test, p=0.04) (Figure 5C and Table 3).
 |
Figure 5 LINC01268 and MAP3K7 gene expressions are linked to drive HCC progression. (A) Positive Pearson coefficient between LINC01268 and MAP3K7 was extracted through GEPIA analysis (http://gepia.cancer-pku.cn) under the correlation mode by choosing liver hepatocellular carcinoma (LIHC) tumor and LIHC normal. High expression of MAP3K7 is related to poor HCC patient overall survival (B), moderate/poor differentiation of HCC (C), and observed in the selected human HCC cell lines (D). Included in the LINC01268 expression assay were a normal human hepatic cell line (LO2) for comparison and three human HCC cell lines (Huh7, HepG2 and Hep3B). The histograms in D showed the mean ± SD of three independent experiments. β-ACTIN served as an internal control in RT-qPCR. *p<0.05, Student’s t-test. |
 |
Table 3 Relationship Between MAP3K7 Expression and Clinical Characteristics of HCC |
Given the elevated MAP3K7 expression levels in cancer tissues of HCC patients (Figure 5C), we determined the abundance of MAP3K7 in three HCC cell lines (Huh7, HepG2 and Hep3B) by RT-qPCR. Similar to the HCC patient biopsies, we found that MAP3K7 expression is elevated (4- to 8-fold) in the three human HCC cell lines, but not in the human fetal hepatocyte cell line LO2 (Figure 5D). Overall, our data suggested that highly expressed LINC01268 and MAP3K7 may play a key role in the progression of HCC and could have prognostic value.
LINC01268 Could Regulate the Expression of MAP3K7
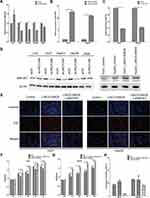
In addition to the data from liver cancer patients and HCC cell lines that indicated synchronistic expression of LINC01268 and MAP3K7 genes (Figure 5A and D), the impact of altered LINC01268 expression on MAP3K7 abundance was tested by RT-qPCR and Western blot. Two days after knockdown of LINC01268, a significant reduction (Student’s t-test, p<0.05) of MAP3K7 expression levels in Huh7 (to 58%), HepG2 (65%) and Hep3B (60%) were observed, when compared to non-targeting siRNA control, with no significant change in LO2 (145%) or A549 (79%) cells (Figure 6A). In addition, we verified that the dependencies of LINC01268 expression was not only residing on the transcriptional level but also reflected on the MAP3K7 protein level (Figure 6D, left panel). These results implied that MAP3K7 expression is, at least in part, determined by LINC01268 in HCC cell lines.
Downregulation of MAP3K7 in HCC Cells Attenuated LINC01268 Overexpression Mediated Increase of Cell Proliferation and Reduction of Cell Apoptosis
On one hand, both LINC01268 and MAPK3K7 are highly expressed in HCC biopsies and cells (Figures 2, 3 and 5), on the other hand, their expression profiles are positively correlated in HCC tissues (Figure 5A), and knockdown of LINC01268 suppressed the expression of MAP3K7 in HCC cell lines at both mRNA (Figure 6A) and protein (Figure 6D, left panel) levels. We therefore speculate that the abnormal high expression of LINC01268 is associated with HCC progression via regulating MAP3K7. In order to validate this hypothesis, we generated stable LINC01268-overexpressing HepG2 and Huh7 cell lines to investigate whether MAP3K7 downregulation would reverse or attenuate the biological phenotypes resulted from LINC01268 overexpression.
Compared to the negative control (transfected with empty vector, LINC01268NC), overexpression of LINC01268 (LINC01268OE) increased the RNA level of LINC01268 by >10 time (Figure 6B) and elevated the protein level of MAP3K7 (Figure 6D, right panel) in both HepG2 and Huh7 cell lines. As expected, opposite to the LINC01268 knockdown, the LINC01268 overexpression (LINC01268OE) led to enhanced proliferation of both HepG2 and Huh7 cells measured by EdU assay after two days cell culture (Figure 6E) and colorimetric measurement over 4 days culture (Figure 6F and G), while upregulation of LINC01268 significantly inhibited cell apoptosis which was measured after 48 h cell culture (Figure 6H). In agreement with our hypothesis, as shown in Figure 6E–H, simultaneous knockdown of MAP3K7 (Figure 6C) in stable LINC01268-overexpressing HepG2 and Huh7 cell lines (denoted as LINC01268OE+siMAP3K7) reversed the increase in cell proliferation and decreased cell apoptosis, to a similar level as observed in the control cells (NC). Taken together, we conclude that the abnormal high expression of LINC01268 is associated with HCC progression via regulating MAP3K7.
Discussion
Hepatocellular carcinoma (HCC) causes high rate of morbidity and mortality. Although early surgery and liver transplantation can prolong the lifespan of patients, the rate of reoccurrence and mortality remains extremely high.50 Identifying specific markers is therefore urgently needed for early diagnosis and prognosis of HCC. Advances in next-generation sequencing technologies has enabled the annotation of tgousands of lncRNAs and many of them have the potential to be prominent cancer markers.51,52 The pathological grade of tumors is one of the most important indicators to evaluate or predict malignant degree and prognosis. Our detailed studies identified LINC01268 to be highly expressed in poorly/moderately differentiated HCC (Figure 2B and D). LINC01268 has putative tumor-promoting function since its high abundance increased HCC cell proliferation (Figure 6E–G) and suppressed cell apoptosis (Figure 6H). Publicly available data41,42 revealed that high expression of LINC01268 was associated with shorter overall survival (Figure 2A), which could be of prognostic value. Since higher LINC01268 expression was more frequent in moderately or poorly differentiated HCC and in HCC with two or more nodules, we suggest that LINC01268 expression contributes to a more localized response and more severe malignancy when the cancer is already established rather than to initiated liver cancer or to promote metastasis. However, at which stage of carcinogenesis and how the expression of LINC01268 is dynamically deregulated requires further investigations. Furthermore, identifying the transcriptional factor(s) that initiate gene expression of LINC01268 can help to further explain the observed expression pattern in HCC.
To further strengthen our observations and findings, the effects of the knockdown of LINC01268 on other tumorigenic properties of cells in vitro and/or in vivo are favorable, for example, in vitro assays of migration and invasion properties would be carried out. Demonstrating the importance of LINC01268 could also be reinforced by a complementary approach upon LINC01268 overexpression.
Upon transcription, many lncRNAs function by forming complexes with proteins and nucleic acids to regulate coding and noncoding genes, transcripts and proteins.53 Since fluorescence in situ hybridization in both HCC tissues and cells indicated nuclear and cytoplasmic localization, we speculate that LINC01268 can exert its functions both at transcriptional and posttranscriptional levels. We observed a stronger positive correlation between LINC01268 and MAP3K7 (Figure 5A, Pearson correlation coefficient, R=0.39, p<0.01). Based on the current human genome annotation (GRCh38.p13), LINC01268 and MAP3K7 are more than 23,000 kb apart and it is therefore unlikely that they influence each other’s gene expression in cis. Interestingly, publicly available genome interaction data (Hi-C) in HepG2 cells54,55 suggests that the MAP3K7 gene locus is located several topologically associated domains (TADs) apart from the LINC01268 gene locus . At present, no apparent genomic connection in trans can be determined. Detailed chromatin confirmation studies along with deleting or replacing LINC01268 gene locus could provide further insight into how LINC01268 interacts with its neighboring genes, as the possibility that LINC01268 encodes functional peptides has been discarded previously.44
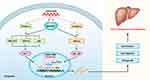
It has been shown that MAP3K7 accelerate HCC tumorigenesis and metastasis via the activation of NF-κB, MAPK and some other pathways.45,47–49 A recent kinome siRNA library screening revealed that the reduction of MAP3K7 expression diminished the growth, migration and invasion of HCC cells.46 Since our data showed that downregulation of LINC01268 reduced the expression of MAP3K7 on the mRNA and protein levels in HCC cells, we proposed a potential mechanism (Figure 7) here speculating that LINC01268 acts on the upstream of the MAP3K7-NFκB-MAPK pathway to drive HCC progression. How LINC01268 and MAP3K7 expressions are linked requires further investigation. For example, the identifications of partners (DNA, RNA or protein) binding to LINC01268 or closer dissection of the genes and signaling pathways, which are affected downstream of LINC01268 and MAP3K7, might provide more detailed information. Further in-depth molecular studies could then also clarify how LINC01268 promotes cell proliferation as we have detected in our study. It is also possible that LINC01268 contributes to HCC cell migration and invasion, which could lead to metastasis and tumorigenesis. Larger HCC cohort studies and detailed clinical information would be necessary to discern the role of LINC01268 in cancer initiation, promotion, progression and metastasis.
In our study we showed that MAP3K7 is an important downstream regulatory target of LINC01268 in HCC, either direct or indirect (via regulating the expression of an intermediate protein or more). We revealed that the knockdown of MAP3K7 attenuated the cellular phenotypes resulted from overexpression of LINC01268 (Figure 6). Alternatively, a demonstration that overexpression of MAP3K7 neutralizes the knockdown of LINC01268 would further support the finding. Future mechanistic studies would allow to dissect the important link between LINC01268 and MAP3K7.
Conclusion
In conclusion, we demonstrated that LINC01268 was highly expressed in HCC cells and tissues in a distinct spatial and temporal fashion. Since high expression of LINC01268 was linked to poor overall survival, LINC01268 could be a novel biomarker for HCC diagnosis, progression and/or prognosis. Our data also suggest MAP3K7 as a downstream effector through which LINC01268 could operate directly or indirectly to promote HCC cell proliferation and differentiation.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Ethics Approval and Consent to Participate
Informed consents were obtained from all patients and all experiments were carried out according to the guidelines of the Declaration of Helsinki and the Ethics Committee of the First Affiliated Hospital of China Medical University (License number 2019-172-4).
Acknowledgments
We would like to thank the staff in the Central Laboratory of China Medical University for the guidance on experimental details. Thanks to the surgical assistants and students of the Department of Hepatobiliary Surgery, the First Affiliated Hospital of China Medical University, for helping collect liver cancer tissue specimens.
Author Contributions
All authors made a significant contribution to this work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all the following areas: took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; agreed on the journal to which the article has been submitted; and agreed to be accountable for all aspects of the work.
Funding
This research was supported by National Natural Science Foundation of China (Grant Number: 81570519).
Disclosure
The authors declare that they have no competing interests.
References
1. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi:10.1038/s41575-019-0186-y
2. Jafri W, Kamran M. Hepatocellular carcinoma in Asia: a challenging situation. Euroasian J Hepatogastroenterol. 2019;9(1):27–33. doi:10.5005/jp-journals-10018-1292
3. Lei Y, Yee LW, Ping ZW. Target patients for partial hepatectomy and relationship between PLT and prognosis in BCLC B HCC. J Hepatol. 2015;62(3):750. doi:10.1016/j.jhep.2014.11.006
4. AlSalloom AA. An update of biochemical markers of hepatocellular carcinoma. Int J Health Sci (Qassim). 2016;10(1):121–136.
5. Zhao YJ, Ju Q, Li GC. Tumor markers for hepatocellular carcinoma. Mol Clin Oncol. 2013;1(4):593–598. doi:10.3892/mco.2013.119
6. Zhao S, Long M, Zhang X, et al. The diagnostic value of the combination of Golgi protein 73, glypican-3 and alpha-fetoprotein in hepatocellular carcinoma: a diagnostic meta-analysis. Ann Transl Med. 2020;8(8):536. doi:10.21037/atm.2020.02.89
7. Lou J, Zhang L, Lv S, Zhang C, Jiang S. Biomarkers for hepatocellular carcinoma. Biomark Cancer. 2017;9:1–9. doi:10.1177/1179299X16684640
8. Consortium ITP-CAoWG. Pan-cancer analysis of whole genomes. Nature. 2020;578(7793):82–93. doi:10.1038/s41586-020-1969-6
9. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi:10.1101/gad.1800909
10. Rafiee A, Riazi-Rad F, Havaskary M, Nuri F. Long noncoding RNAs: regulation, function and cancer. Biotechnol Genet Eng Rev. 2018;34(2):153–180. doi:10.1080/02648725.2018.1471566
11. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi:10.1016/j.ccell.2016.03.010
12. Ransohoff JD, Wei YN, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Bio. 2018;19(3):143–157. doi:10.1038/nrm.2017.104
13. Huang JL, Zheng L, Hu YW, Wang Q. Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis. 2014;35(3):507–514. doi:10.1093/carcin/bgt405
14. Quagliata L, Matter MS, Piscuoglio S, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59(3):911–923. doi:10.1002/hep.26740
15. Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. doi:10.1016/j.ccr.2014.03.010
16. Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int. 2013;2013:136106. doi:10.1155/2013/136106
17. Panzitt K, Tschernatsch MM, Guelly C, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132(1):330–342. doi:10.1053/j.gastro.2006.08.026
18. Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38(16):5366–5383. doi:10.1093/nar/gkq285
19. Du Y, Kong G, You X, et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287(31):26302–26311. doi:10.1074/jbc.M112.342113
20. Maass PG, Luft FC, Bahring S. Long non-coding RNA in health and disease. J Mol Med (Berl). 2014;92(4):337–346. doi:10.1007/s00109-014-1131-8
21. Li H, An J, Wu M, et al. LncRNA HOTAIR promotes human liver cancer stem cell malignant growth through downregulation of SETD2. Oncotarget. 2015;6(29):27847–27864. doi:10.18632/oncotarget.4443
22. Pu H, Zheng Q, Li H, et al. CUDR promotes liver cancer stem cell growth through upregulating TERT and C-Myc. Oncotarget. 2015;6(38):40775–40798. doi:10.18632/oncotarget.5805
23. Wang Y, He L, Du Y, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16(4):413–425. doi:10.1016/j.stem.2015.03.003
24. Mello SS, Sinow C, Raj N, et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017;31(11):1095–1108. doi:10.1101/gad.284661.116
25. Chakravarty D, Sboner A, Nair SS, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi:10.1038/ncomms6383
26. Adriaens C, Standaert L, Barra J, et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22(8):861–868. doi:10.1038/nm.4135
27. Yu W, Qiao Y, Tang X, et al. Tumor suppressor long non-coding RNA, MT1DP is negatively regulated by YAP and Runx2 to inhibit FoxA1 in liver cancer cells. Cell Signal. 2014;26(12):2961–2968. doi:10.1016/j.cellsig.2014.09.011
28. Ding LJ, Li Y, Wang SD, et al. Long Noncoding RNA lncCAMTA1 promotes proliferation and cancer stem cell-like properties of liver cancer by inhibiting CAMTA1. Int J Mol Sci. 2016;17:10. doi:10.3390/ijms17101617
29. Wang X, Sun W, Shen W, et al. Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J Hepatol. 2016;64(6):1283–1294. doi:10.1016/j.jhep.2016.01.019
30. Lanzafame M, Bianco G, Terracciano LM, Ng CKY, Piscuoglio S. The role of long non-coding RNAs in hepatocarcinogenesis. Int J Mol Sci. 2018;19:3. doi:10.3390/ijms19030682
31. El Khodiry A, Afify M, El Tayebi HM. Behind the curtain of non-coding RNAs; long non-coding RNAs regulating hepatocarcinogenesis. World J Gastroenterol. 2018;24(5):549–572. doi:10.3748/wjg.v24.i5.549
32. Punzi G, Ursini G, Viscanti G, et al. Association of a noncoding RNA postmortem with suicide by violent means and in vivo with aggressive phenotypes. Biol Psychiatry. 2019;85(5):417–424. doi:10.1016/j.biopsych.2018.11.002
33. Punzi G, Ursini G, Shin JH, Kleinman JE, Hyde TM, Weinberger DR. Increased expression of MARCKS in post-mortem brain of violent suicide completers is related to transcription of a long, noncoding, antisense RNA. Mol Psychiatry. 2014;19(10):1057–1059. doi:10.1038/mp.2014.41
34. Matjasic A, Popovic M, Matos B, Glavac D. Expression of LOC285758, a potential long non-coding biomarker, is methylation-dependent and correlates with glioma malignancy grade. Radiol Oncol. 2017;51(3):331–341. doi:10.1515/raon-2017-0004
35. Lei L, Xia S, Liu D, et al. Genome-wide characterization of lncRNAs in acute myeloid leukemia. Brief Bioinform. 2018;19(4):627–635. doi:10.1093/bib/bbx007
36. Luedde T, Schwabe RF. NF-kappaB in the liver–linking injury, fibrosis and hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2011;8(2):108–118. doi:10.1038/nrgastro.2010.213
37. Wang SN, Lee KT, Tsai CJ, Chen YJ, Yeh YT. Phosphorylated p38 and JNK MAPK proteins in hepatocellular carcinoma. Eur J Clin Invest. 2012;42(12):1295–1301. doi:10.1111/eci.12003
38. Yang S, Liu G. Targeting the Ras/Raf/MEK/ERK pathway in hepatocellular carcinoma. Oncol Lett. 2017;13(3):1041–1047. doi:10.3892/ol.2017.5557
39. Yang C, Cao M, Liu Y, et al. Inducible formation of leader cells driven by CD44 switching gives rise to collective invasion and metastases in luminal breast carcinomas. Oncogene. 2019;38(46):7113–7132. doi:10.1038/s41388-019-0899-y
40. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
41. Cancer Genome Atlas Research N; Weinstein JN, Collisson EA, Mills GB, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi:10.1038/ng.2764.
42. Consortium GT, Thomas J, Salvatore M. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi:10.1038/ng.2653
43. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi:10.1038/nrg2521
44. Zhang Q, Chao TC, Patil VS, et al. The long noncoding RNA ROCKI regulates inflammatory gene expression. EMBO J. 2019;38:8. doi:10.15252/embj.2018100041
45. Greten FR. TAK1: another mesh in the NF-kappaB - JNK controlled network causing hepatocellular carcinoma. J Hepatol. 2011;55(3):721–723. doi:10.1016/j.jhep.2011.02.037
46. Cheng JS, Tsai WL, Liu PF, et al. The MAP3K7-mTOR axis promotes the proliferation and malignancy of hepatocellular carcinoma cells. Front Oncol. 2019;9:474. doi:10.3389/fonc.2019.00474
47. Wang Y, Tu Q, Yan W, et al. CXC195 suppresses proliferation and inflammatory response in LPS-induced human hepatocellular carcinoma cells via regulating TLR4-MyD88-TAK1-mediated NF-kappaB and MAPK pathway. Biochem Biophys Res Commun. 2015;456(1):373–379. doi:10.1016/j.bbrc.2014.11.090
48. Zhao N, Wang R, Zhou L, Zhu Y, Gong J, Zhuang SM. MicroRNA-26b suppresses the NF-kappaB signaling and enhances the chemosensitivity of hepatocellular carcinoma cells by targeting TAK1 and TAB3. Mol Cancer. 2014;13:35. doi:10.1186/1476-4598-13-35
49. Tey SK, Tse EYT, Mao X, et al. Nuclear Met promotes hepatocellular carcinoma tumorigenesis and metastasis by upregulation of TAK1 and activation of NF-kappaB pathway. Cancer Lett. 2017;411:150–161. doi:10.1016/j.canlet.2017.09.047
50. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. doi:10.1016/S0140-6736(18)30010-2
51. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi:10.1038/nm.3981
52. Li C, Chen J, Zhang K, Feng B, Wang R, Chen L. Progress and Prospects of Long Noncoding RNAs (lncRNAs) in hepatocellular carcinoma. Cell Physiol Biochem. 2015;36(2):423–434. doi:10.1159/000430109
53. Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20(3):300–307. doi:10.1038/nsmb.2480
54. Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74.
55. Wang Y, Song F, Zhang B, et al. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018;19(1):151. doi:10.1186/s13059-018-1519-9
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.