Back to Journals » Journal of Inflammation Research » Volume 15
High Expression of DEPDC1B Predicts Poor Prognosis in Lung Adenocarcinoma
Authors Li P, Chen X, Zhou S, Xia X, Wang E, Han R, Zeng D, Fei G, Wang R
Received 3 April 2022
Accepted for publication 11 July 2022
Published 23 July 2022 Volume 2022:15 Pages 4171—4184
DOI https://doi.org/10.2147/JIR.S369219
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Monika Sharma
Pulin Li,1,* Xiaojuan Chen,2,* Sijing Zhou,3,* Xingyuan Xia,1 Enze Wang,1 Rui Han,1 Daxiong Zeng,4,5 Guanghe Fei,1 Ran Wang1
1Department of Respiratory and Critical Care Medicine, the First Affiliated Hospital of Anhui Medical University, Hefei, People’s Republic of China; 2Department of Infectious Diseases, Hefei Second People’s Hospital, Hefei, People’s Republic of China; 3Department of Occupational Medicine, Hefei Third Clinical College of Anhui Medical University, Hefei, People’s Republic of China; 4Department of Pulmonary and Critical Care Medicine, Suzhou Dushu Lake Hospital, Suzhou, People’s Republic of China; 5Department of Pulmonary and Critical Care Medicine, Dushu Lake Hospital Affiliated to Soochow University, Medical Center of Soochow University, Suzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Guanghe Fei; Ran Wang, Department of Respiratory and Critical Care Medicine, the First Affiliated Hospital of Anhui Medical University, Hefei, People’s Republic of China, Tel/Fax +86 -551-62922913, Email [email protected]; [email protected]
Introduction: Lung adenocarcinoma (LUAD) is the most common type of lung cancer. DEP domain-containing 1 B (DEPDC1B) is involved in the development of several cancers; however, its role in LUAD is unknown. Therefore, we aimed to determine the biological function and prognostic value of DEPDC1B in LUAD.
Material and Methods: We analyzed the correlation between DEPDC1B expression and the clinical features of LUAD and lung squamous cell carcinoma (LUSC). Survival was evaluated by generating Kaplan–Meier curves, which were used to analyze the relationship between DEPDC1B expression and prognosis in LUAD and LUSC. DEPDC1B expression in tumor and normal tissues from patients with LUAD and LUSC was determined using immunohistochemistry, and its clinical significance was analyzed. Finally, the correlation between the expression and biological function of DEPDC1B in LUAD was examined.
Results: Our findings revealed that DEPDC1B expression was higher in tumor tissues than that in normal tissues from patients with LUAD and LUSC (P < 0.001). These results were confirmed in clinical samples from patients using immunohistochemistry. Analysis of a dataset from The Cancer Genome Atlas (TCGA) showed that high DEPDC1B expression was associated with poor prognosis only in patients with LUAD (P < 0.001). Similarly, high DEPDC1B expression was related to shorter overall survival (OS) and progression-free interval (PFI) in patients with LUAD. These associations were not observed in LUSC. Functional enrichment analysis suggested that DEPDC1B promoted tumor development in LUAD by regulating the cell cycle.
Conclusion: High DEPDC1B expression predicts poor prognosis in patients with LUAD. Thus, DEPDC1B has potential as a therapeutic target for LUAD.
Keywords: DEP domain-containing 1B, lung adenocarcinoma, prognosis, immunohistochemistry, bioinformatics
Graphical Abstract:
Introduction
Lung cancer is a leading cause of cancer and cancer-related death worldwide.1–3 The majority of lung cancers (about 85%) are non-small cell lung cancer (NSCLC), which includes lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). Adenocarcinoma is the most common pathological subtype. Prior to the 1990s, LUSC was the most common pathological subtype, especially in males. However, the incidence of adenocarcinoma has increased and now exceeds that of squamous cell carcinoma.4 Lung cancer is one of the deadliest cancers, accounting for approximately 28% of cancer-related deaths. Most lung cancers are diagnosed at advanced stages, and the five-year survival rate is poor.5 Despite recent advances in the timely diagnosis and treatment of NSCLC, the prognosis of advanced NSCLC remains poor. The diagnosis and treatment of NSCLC are multidisciplinary and require comprehensive clinical, radiological, molecular biological, and pathological data to accurately assess the prognosis and identify therapeutic targets. Studies have shown that molecular biological detection plays a significant role in improving the early diagnosis rate and prognosis of NSCLC.6–8 Therefore, identifying oncogenes associated with prognosis in NSCLC is important for developing molecular biomarkers.
DEP domain-containing 1 B (DEPDC1B) is involved in various signaling pathways. DEPDC1B, located on chromosome 5q12, encodes a 61 kDa protein of 529 amino acids.9,10 DEP domains play significant roles in several processes. DEPDC1B recognizes G-protein-coupled receptors and regulates signaling pathways through effector and regulatory factors.11 DEPDC1B interacts with diverse signaling molecules, including splicing regulatory molecules and transmembrane proteins. Studies have shown that DEPDC1B participates in cell adhesion, cell proliferation, and cell cycle regulation.12,13 DEPDC1B is overexpressed in various cancers and is a potential biomarker and therapeutic target.9,14–18 However, few studies have explored the role of DEPDC1B in NSCLC. Therefore, the objective of the present study was to explore the correlation between DEPDC1B expression and the clinicopathological features and prognosis of NSCLC. We also aimed to clarify the biological processes involving DEPDC1B in LUAD.
We analyzed and compared DEPDC1B expression between tumor and normal tissues in LUAD and LUSC in dataset from The Cancer Genome Atlas (TCGA) database. We then assessed DEPDC1B expression in clinical LUAD and LUSC samples using immunohistochemistry. We separately validated the association between DEPDC1B expression and prognosis in patients with LUAD and LUSC. Then, we used gene set enrichment analysis (GSEA) to determine the possible functions of DEPDC1B in LUAD. Our results strongly support DEPDC1B as a biomarker for predicting prognosis and treatment outcomes in LUAD. GSEA and immune-associated infiltration analysis revealed the biological impact of DEPDC1B in LUAD and the underlying mechanism. Our findings indicate that DEPDC1B may be an useful biomarker for predicting the prognosis of LUAD, but not LUSC.
Materials and Methods
Bioinformatics Data Source
We collected 594 LUAD cases (535 tumors and 59 normal tissues) and 551 LUSC cases (502 tumors and 49 normal tissues) from TCGA (https://cancergenome.nih.gov; Table S1). Next, we divided the cases into two groups, low and high DEPDC1B expression groups, using the median expression level as a cutoff.
Pathological Sample Collection
We collected tumor tissue samples and paired adjacent normal tissues (normal lung tissue more than 5 cm from the tumor margin) from 100 patients diagnosed with LUAD and 60 patients diagnosed with LUSC between December 2017 and December 2020. All patients were admitted to the First Affiliated Hospital of Anhui Medical University and provided written informed consent. The follow-up date was December 31, 2021 or the date of death. We collected the following patient information: age, gender, smoking status, pathological stage, TNM stage, tumor differentiation, follow-up status, progression free interval (PFI), and overall survival (OS). The clinicopathological information for enrolled patients is presented in Table S2. Two pathologists diagnosed lung cancer based on pathological findings and determined histologic types and stages according to the eighth edition of the tumor, node, metastasis (TNM) staging system for lung cancer.19 This study was approved by the Institutional Review Board of First Affiliated Hospital of Anhui Medical University.
Immunohistochemical Analysis of DEPDC1B Expression
DEPDC1B expression in LUAD and LUSC was evaluated through immunohistochemical analysis of tumor and normal tissue samples collected from 100 patients diagnosed with LUAD and 60 patients diagnosed with LUSC. The tissues were fixed with formalin and embedded in paraffin, and DEPDC1B was detected using a rabbit anti- DEPDC1B polyclonal antibody (ab237542; Abcam). The paraffin embedded slides were deparaffinized in xylene, dehydrated using an ethanol gradient, and rinsed with distilled water and phosphate-buffered saline. Antigens were extracted by incubation in EDTA buffer for 30 min. Sections were incubated with an anti-DEPDC1B antibody (1:100 dilution) overnight at 4°C and then incubated with HRP-conjugated goat anti-rabbit IgG (ab111909; Abcam) at room temperature for 2 h. Finally, the slides were developed with DAB, stained with hematoxylin, dehydrated, mounted, examined, and evaluated. Two independent pathologists examined the prepared slides under a light microscope (400×) along with positive and negative controls. DEPDC1B overexpression was calculated based on the percentage of stained cells and staining intensity using a semi-quantitative scoring system.20 Scores were assigned according to the percentage of stained cells as follows: 0 (0–5%), 1 (5–25%), 2 (25–50%), 3 (50–75%), and 4 (75–100%), and staining intensity was scored according to the color of the cell as follows: 0 (no staining), 1 (light staining), 2 (moderate staining), or 3 (intense staining). The scores for the percentage of stained cells and staining intensity were multiplied to obtain the final score (0–12 points). We defined 0–2, 3–6, and 7–12 points as no expression, low DEPDC1B expression, and high DEPDC1B expression, respectively.
Differential Gene Expression Analysis
We divided patients with LUAD into low and high DEPDC1B expression groups using the median z-score. Differentially expressed genes (DEGs) were analyzed using R software with a |log fold change| (log FC) >1 and adjusted P value <0.05 as threshold values.21 Volcano and heat maps were created to visualize the results of the analyses.
Gene Set Enrichment Analysis
The biological function of DEPDC1B in LUAD was explored using Gene Set Enrichment Analysis (GSEA) of Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. GSEA has the advantage of gene sets grouped based on common biological function, chromosomal regulatory mechanism, or location.22 Enrichment results were considered statistically significant when they showed a false discovery rate (FDR) <0.25 and an adjusted P-value <0.05.
Immune Infiltrate Analysis
Immune infiltration in LUAD samples was analyzed by single-sample GSEA, and infiltration of 24 immune cell types was analyzed using the GSVA package in R (http://www.biocondutor.org/package/release/bioc/html/GSVA.html).23
Statistical Analysis
We analyzed DEPDC1B expression in unpaired and paired samples using the Wilcoxon rank-sum and Wilcoxon rank-sum sign tests, respectively. A nomogram of the clinical variables associated with DEPDC1B in LUAD was generated using the RMS R package.24 The prognostic value of DEPDC1B in LUAD and LUSC was assessed using Kaplan–Meier analysis. Univariate and multivariate Cox regression analyses were employed to determine the prognostic factors in LUAD. All statistical analyses were conducted and plots were generated using R software (version 3.6.3). A P-value less than 0.05 was considered statistically significant.
Results
Elevated Expression of DEPDC1B in LUAD and LUSC
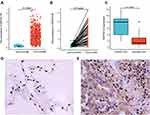
To examine DEPDC1B expression in patients with LUAD and LUSC, we compared the expression levels in tumor and normal tissues in a dataset from TCGA. In LUAD, DEPDC1B levels were significantly higher in tumor tissues than in normal tissues (P < 0.001; Figure 1A) and were higher in tumor tissues than in paired normal tissues (P < 0.001; Figure 1B). Similar results were observed in LUSC (P < 0.001; Figure S1A and B). To verify this difference in DEPDC1B expression, immunohistochemical analysis was performed using paired normal and tumor tissues collected from 100 patients with LUAD and 60 patients with LUSC. The results of our analysis confirmed elevated DEPDC1B levels in LUAD tumor tissue (P < 0.001; Figure 1C), and representative immunohistochemical images are shown in Figure 1D and E. Similar results were observed in LUSC (P < 0.001; Figure S1C–E).
Relationship Between DEPDC1B Expression and the Clinicopathologic Features of LUAD and LUSC Obtained from a TCGA Dataset
Kruskal–Wallis analysis showed that DEPDC1B expression levels in patients with LUAD were significantly related to T stage (P < 0.001), N stage (P = 0.046), primary therapy outcome (P = 0.004), and pathologic stage (P = 0.028; Figure 2A–D). DEPDC1B expression levels in patients with LUSC were significantly related to T stage (P = 0.042), N stage (P < 0.001), and pathologic stage (P< 0.001) (Figure S2A–C). Results of the Mann–Whitney U-test revealed that DEPDC1B levels were significantly related to gender (P = 0.003), age (P = 0.025), and smoker (P = 0.048; Figure 2E–G) in LUAD, but only significantly related to age (P = 0.003; Figure S2D) in LUSC.
High Expression of DEPDC1B is Associated with Poor Prognosis in LUAD
Kaplan–Meier survival analysis of TCGA dataset showed that patients with high DEPDC1B-expressing tumors had shorter survival than those with low DEPDC1B-expressing tumors (OS, Figure 3A and PFI, Figure 3B; P < 0.001). These results were validated in 100 patients with LUAD (OS, Figure 3C and PFI, Figure 3D; P < 0.001). Interestingly, no significant difference was observed in patients with LUSC in TCGA dataset (Figure S3A and B) or our 60 LUSC patient cohort (Figure S3C and D). Therefore, we only conducted subgroup analysis of patients with LUAD in TCGA dataset, which showed that high DEPDC1B expression was correlated with OS for different TNM stages: T1 and T2 (P = 0.002; Figure S4A), T3 and T4 (P = 0.016, Figure S4B); N0 (P = 0.021; Figure S4C), N1, N2, and N3 (P = 0.065; Figure S4D); M0 (P < 0.001; Figure S4E) and M1 (P = 0.75; Figure S4F). Similar results were observed for clinical stages (I and II, P = 0.002, Figure S4G; III and IV, P = 0.116, Figure S4H). These results suggest that DEPDC1B expression levels impact the prognosis of patients with LUAD (Figure 4A).
Univariate and Multivariate Cox Analyses of Prognosis in Patients with LUAD
We examined the correlation between DEPDC1B expression and clinical characteristics of patients with LUAD in TCGA dataset using univariate Cox analysis. Clinicopathological variables associated with shorter survival included TNM stage, pathologic stage, primary therapy outcome, and high DEPDC1B expression. To comprehensively explore the factors related to high DEPDC1B expression and survival, multivariate Cox regression analysis was performed; high DEPDC1B expression remained an independent factor correlated with OS (Hazard ratio [HR]: 1.949, 95% confidence interval [CI]: 1.292–2.940; P = 0.001) and was correlated with N stage and primary therapy outcome (Table 1). We also conducted univariate and multivariate Cox analyses of our 100-patient cohort with LUAD. Univariate analysis showed that gender, N stage, pathologic stage, differentiation, and DEPDC1B expression were prognostic factors for OS. The results of the univariate analysis of PFI were similar to those of OS and are shown in Table 2. Multivariate analysis was performed using variables with P values <0.05 in the univariate analysis, and the results showed that male sex, terminal pathologic stage, poor differentiation, and high DEPDC1B expression were independent risk factors for OS and PFI of patients with LUAD, and the detailed results are shown in Table 2.
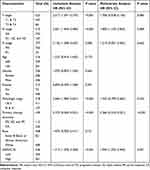 |
Table 1 Correlations Between DEPDC1B Expression and Overall Survival in Patients with Lung Adenocarcinoma Based on Univariate and Multivariate Cox Regression Analysis |
 |
Table 2 Univariate and Multivariate Analyses of Overall Survival and Progression Free Interval in 100 Patients with Lung Adenocarcinoma |
Construction of a Nomogram Based on DEPDC1B Expression in LUAD
To better predict the outcomes of patients with LUAD, we constructed a nomogram using the multivariate analysis of TCGA dataset. This line chart incorporates the clinical features independently associated with survival in the multivariate analysis (ie, TNM stage, primary therapy outcome, and DEPDC1B expression; Figure 4B). High DEPDC1B expression was associated with poor prognosis, in agreement with the results of the multivariate analysis. The c-index of the line chart was 0.719 (95% CI: 0.693–0.745). The calibration plot (Figure S5) showed that the predicted values are in good agreement with the observed values.
Identification of DEGs Between LUAD Patients with Low and High DEPDC1B Expression
We used R (with |logFC| >1, and an adjusted P value <0.05) to analyze TCGA dataset (Figure 5). We identified 3533 DEGs (2325 upregulated and 1208 downregulated) between the low and high DEPDC1B expression groups. Volcano and heat maps were created to visualize the results (Figure 5A and B).
DEPDC1B-Related Signaling Pathways and Enrichment Analyses in LUAD
To elucidate the role of DEPDC1B in LUAD, we conducted GO enrichment analysis of the 1400 DEGs most closely related to DEPDC1B using the ClusterProfile R package. We detected 253 enriched terms in GO biological process categories, including “organelle fission”, “nuclear division”, and “chromosome segregation” (Figure 5C), which indicates that DEPDC1B expression is involved in diverse biological processes. Sixty-four GO terms were related to the cellular component category “chromosomal region” (Figure 5D). Furthermore, the analysis suggested enrichment in the molecular function category “receptor-ligand activities” (Figure 5E).
Related signaling pathways were identified in the low and high DEPDC1B expression groups using GSEA, and results with an adjusted P value <0.05 and a FDR <0.25 were regarded as significant. The following eight pathways were significantly different between the two groups: retinoblastoma gene in cancer, cell cycle checkpoints, PLK1 pathway, mitotic spindle checkpoint, deposition of new CENP-A-containing nucleosomes at the centromere, meiotic recombination, DNA replication, and resolution of sister chromatid cohesion (Figure 6A–H).
Correlations Between Immune Cell Infiltration and DEPDC1B Expression
We next explored the correlation between DEPDC1B expression levels and infiltration of 24 immune cell types in LUAD (Figure 7A). DEPDC1B expression was negatively associated with mast cells, eosinophils, immature dendritic cells (iDCs), follicular helper T cells (TFHs), dendritic cells (DCs), CD8 T cells, and natural killer cells (Figure 7B–E) and was positively associated with type-2 T helper (Th2) cells and gamma delta T (Tgd) cells (Figure 7F and G).
Discussion
Lung cancer is one of the deadliest cancers worldwide, and LUAD is the most common histological subtype.2 Developments in chemotherapy and advances in targeted therapy and immunotherapy have led considerable progress in the treatment of LUAD. However, the 5-year survival rate of LUAD remains unsatisfactory.25,26 Owing to the insidious symptoms of early lung cancers, many patients are diagnosed at an advanced stage. Therefore, identifying molecular biomarkers of LUAD is critical to develop novel therapeutic targets and establish optical treatment strategies.
The DEPDC protein family comprises seven members, which are characterized by the presence of a DEP domain. Accumulating evidence indicates that the DEPDC protein family is involved in the development and progression of several cancers, especially hepatocellular carcinoma, and is closely related to poor prognosis.27 DEPDC1B, a member of the DEPDC1 family, is involved in the growth and progression of numerous tumors.9,15–17,27–31
We performed bioinformatics analyses using RNA-seq data from TCGA to assess the prognostic value of DEPDC1B for LUAD and LUSC. The results indicated that high DEPDC1B expression was related to poor OS and PFI in patients with LUAD, but this association was not observed in LUSC. This result was also confirmed using immunohistochemistry. Overexpression of DEPDC1B in LUAD was associated with poor clinicopathological factors, indicating that DEPDC1B may function as an oncogene in LUAD. Typically, LUAD patients with high DEPDC1B expression had poorer OS than patients with low DEPDC1B expression, while no significant difference related to DEPDC1B expression was observed in LUSC. This finding differs from a previous study showing that DEPDC1B acts on the Wnt/β-catenin signaling pathway to accelerate NSCLC migration and invasion, suggesting that elevated DEPDC1B expression may be associated with poor prognosis in both types of NSCLC.32 Previous investigations have indicated that most tumors are related to activation of the Wnt/β-catenin signaling pathway, which is involved in major developmental processes and may promote tumor development by influencing immune exclusion and immunosurveillance.33,34 Therefore, the Wnt/β-catenin signaling may also play an important role in NSCLC. Although DEPDC1B was previously shown to have an oncogenic effect on the progression of NSCLC, the sample size in the study was relatively small, and the possible biological functions of DEPDC1B in LUAD and LUSC have not been comprehensively explored. The results of our study indicate that DEPDC1B plays a role in LUAD but has no significant impact on the prognosis of LUSC. Accordingly, DEPDC1B is more valuable for the prognosis of LUAD.
Given that DEPDC1B expression did not significantly influence the prognosis of patients with LUSC in the present study, functional enrichment and immune infiltration analyses were performed for LUAD only. Targeted therapies for LUAD and LUSC differ substantially;35 targeted therapies for LUAD tend to be more sensitive than those for LUSC. Enrichment analysis suggests that DEPDC1B expression is related to various biological processes, such as mitosis, and plays a significant role in the cell cycle. Studies have also shown that DEPDC1B acts at G2 phase of the cell cycle to control the progression of mitosis through adhesion-dependent signaling mechanisms.12 Sustained proliferative signaling is one of the most important features of tumors.36 Cancer cells are unstable, and our results suggest that high DEPDC1B expression may accelerate mitosis and promote cell migration and proliferation, resulting in tumor development, consistent with previous reports on other cancers.16,31
Evasion of immune destruction is a well-known characteristic of tumors, and immune cell infiltration is an important feature of the tumor microenvironment. The composition and distribution of infiltrating immune cells can impact prognosis. We investigated the relationship between DEPDC1B expression and immune cell infiltration in patients with LUAD. Our results showed that DEPDC1B expression was positively correlated with the levels of Th2 and Tgd cell infiltration. Studies have shown that T-cell stalling dominates the lung cancer landscape, along with immune dysregulation, such as Th1/Th2 cytokine ratio imbalance and T-cell infiltration, leading to tumor invasion.37–39 A previous report showed that a Th2 response and tumor immune evasion lead to poor prognosis; however, this study was conducted in mice, and no significant progress has been made in human studies. Conversely, the development of lung cancer was associated with the expansion and phenotypic changes of resident Tgd cells in the tumor microenvironment.40 In different cancers, Tgd cells regulate tumor activity through distinct mechanisms.41,42 Our findings revealed that DEPDC1B expression in LUAD was negatively associated with infiltration of mast cells and eosinophils. However, infiltration of eosinophils and mast cells has been correlated with a more favorable prognosis in several malignancies.43–45 These results suggest that DEPDC1B might influence immune infiltration to alter the prognosis of patients with LUAD.
Our study has several limitations. First, the study included only one dataset and 160 clinical samples. Thus, the sample size was relatively small, and studies of larger samples are needed to verify the results. Second, although the relationship between DEPDC1B expression and immune cell infiltration was examined using bioinformatics, the specific mechanism requires experimental verification.
Conclusion
Our results showed that the DEPDC1B expression was significantly higher in tumor tissues than in normal tissues in both LUAD and LUSC. We examined the prognostic value of DEPDC1B in LUAD and LUSC, and the results suggest that high DEPDC1B expression predicts poor prognosis in patients with LUAD but not in patients with LUSC. DEPDC1B has potential as a biomarker for predicting the occurrence, progression, and prognosis of LUAD. However, additional experiments are needed to clarify the biological function of DEPDC1B and the underlying mechanism.
Data Sharing Statement
The data of this manuscript can be downloaded from The Cancer Genome Atlas database (https://portal.gdc.cancer.gov/). The original contributions presented in the study are included in the article and supplements. Further inquiries can be directed to the corresponding authors.
Ethics Approval and Consent to Participate
This research had been approved by the Institutional Review Boards of First Affiliated Hospital of Anhui Medical University. This project was following the principles of the Helsinki Declaration.
Consent for Publication
All patients signed an informed consent approved by the Institutional Review Board.
Acknowledgments
We acknowledge the TCGA database for providing their platforms and contributors for uploading their meaningful datasets.
Funding
This research was supported by the fund for Natural Science Foundation of China (No.81970051), Excellent Top Talent Cultivation Project of Anhui Higher Education Institutions (gxgwfx2021014), Construction Project of Provincial Teaching Demonstration Course of Anhui Province (1364), Quality Engineering Project of Anhui medical university, scientific research fund from Anhui medical university (2020xkj257), and the Applied Medical Research Project of Hefei Health Commission (Hwk2021zd008).
Disclosure
The authors have declared that no competing interest exists.
References
1. Ettinger DS, Wood DE, Aisner DL, et al. Non-small cell lung cancer, version 5.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Netw. 2017;15(4):504–535. doi:10.6004/jnccn.2017.0050
2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi:10.3322/caac.21654
3. Zhu K, Xu A, Xia W, et al. Association between NAT2 polymorphism and lung cancer risk: a systematic review and meta-analysis. Front Oncol. 2021;11:567762. doi:10.3389/fonc.2021.567762
4. Lortet-Tieulent J, Soerjomataram I, Ferlay J, Rutherford M, Weiderpass E, Bray F. International trends in lung cancer incidence by histological subtype: adenocarcinoma stabilizing in men but still increasing in women. Lung Cancer. 2014;84(1):13–22. doi:10.1016/j.lungcan.2014.01.009
5. Cheng TY, Cramb SM, Baade PD, Youlden DR, Nwogu C, Reid ME. The international epidemiology of lung cancer: latest trends, disparities, and tumor characteristics. J Thorac Oncol. 2016;11(10):1653–1671. doi:10.1016/j.jtho.2016.05.021
6. Ettinger DS, Wood DE, Aggarwal C, et al. NCCN guidelines insights: non-small cell lung cancer, version 1.2020. J Natl Compr Cancer Netw. 2019;17(12):1464–1472. doi:10.6004/jnccn.2019.0059
7. Vargas AJ, Harris CC. Biomarker development in the precision medicine era: lung cancer as a case study. Nature reviews. Cancer. 2016;16(8):525–537. doi:10.1038/nrc.2016.56
8. Zhang B, Xu A, Wu D, et al. ARL14 as a prognostic biomarker in non-small cell lung cancer. J Inflamm Res. 2021;14:6557–6574. doi:10.2147/jir.S340119
9. Liu X, Li T, Huang X, et al. DEPDC1B promotes migration and invasion in pancreatic ductal adenocarcinoma by activating the Akt/GSK3β/Snail pathway. Oncol Lett. 2020;20(5):146. doi:10.3892/ol.2020.12009
10. Garcia-Mata R. Arrested detachment: a DEPDC1B-mediated de-adhesion mitotic checkpoint. Dev Cell. 2014;31(4):387–389. doi:10.1016/j.devcel.2014.11.008
11. Ballon DR, Flanary PL, Gladue DP, Konopka JB, Dohlman HG, Thorner J. DEP-domain-mediated regulation of GPCR signaling responses. Cell. 2006;126(6):1079–1093. doi:10.1016/j.cell.2006.07.030
12. Marchesi S, Montani F, Deflorian G, et al. DEPDC1B coordinates de-adhesion events and cell-cycle progression at mitosis. Dev Cell. 2014;31(4):420–433. doi:10.1016/j.devcel.2014.09.009
13. Chen D, Ito S, Hyodo T, Asano-Inami E, Yuan H, Senga T. Phosphorylation of DEPDC1 at Ser110 is required to maintain centrosome organization during mitosis. Exp Cell Res. 2017;358(2):101–110. doi:10.1016/j.yexcr.2017.06.005
14. Bai S, Chen T, Du T, et al. High levels of DEPDC1B predict shorter biochemical recurrence-free survival of patients with prostate cancer. Oncol Lett. 2017;14(6):6801–6808. doi:10.3892/ol.2017.7027
15. Chen X, Guo ZQ, Cao D, Chen Y, Chen J. Knockdown of DEPDC1B inhibits the development of glioblastoma. Cancer Cell Int. 2020;20:310. doi:10.1186/s12935-020-01404-7
16. Dang XW, Pan Q, Lin ZH, et al. Overexpressed DEPDC1B contributes to the progression of hepatocellular carcinoma by CDK1. Aging. 2021;13(16):20094–20115. doi:10.18632/aging.203016
17. Lai CH, Xu K, Zhou J, et al. DEPDC1B is a tumor promotor in development of bladder cancer through targeting SHC1. Cell Death Dis. 2020;11(11):986. doi:10.1038/s41419-020-03190-6
18. Zhang S, Shi W, Hu W, et al. DEP domain-containing protein 1B (DEPDC1B) promotes migration and invasion in pancreatic cancer through the Rac1/PAK1-LIMK1-Cofilin1 signaling pathway. Onco Targets Ther. 2020;13:1481–1496. doi:10.2147/ott.S229055
19. Rami-Porta R, Bolejack V, Crowley J, et al. The IASLC lung cancer staging project: proposals for the revisions of the T descriptors in the forthcoming eighth edition of the TNM classification for lung cancer. J Thorac Oncol. 2015;10(7):990–1003. doi:10.1097/jto.0000000000000559
20. Li Q, Wu J, Wei P, et al. Overexpression of forkhead Box C2 promotes tumor metastasis and indicates poor prognosis in colon cancer via regulating epithelial-mesenchymal transition. Am J Cancer Res. 2015;5(6):2022–2034.
21. Dalman MR, Deeter A, Nimishakavi G, Duan ZH. Fold change and p-value cutoffs significantly alter microarray interpretations. BMC Bioinform. 2012;13(Suppl 2):S11. doi:10.1186/1471-2105-13-s2-s11
22. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
23. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi:10.1186/1471-2105-14-7
24. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77. doi:10.1186/1471-2105-12-77
25. Jiang H, Xu A, Xia W, et al. Nivolumab monotherapy or combination therapy with ipilimumab for lung cancer: a systemic review and meta-analysis. Cancer Cell Int. 2021;21(1):426. doi:10.1186/s12935-021-02100-w
26. Zhang B, Liu Y, Zhou S, Jiang H, Zhu K, Wang R. Predictive effect of PD-L1 expression for immune checkpoint inhibitor (PD-1/PD-L1 inhibitors) treatment for non-small cell lung cancer: a meta-analysis. Int Immunopharmacol. 2020;80:106214. doi:10.1016/j.intimp.2020.106214
27. Liao Z, Wang X, Zeng Y, Zou Q. Identification of DEP domain-containing proteins by a machine learning method and experimental analysis of their expression in human HCC tissues. Sci Rep. 2016;6:39655. doi:10.1038/srep39655
28. Ahuja P, Singh K. In Silico Approach for SAR analysis of the predicted model of DEPDC1B: a novel target for oral cancer. Adv Bioinformatics. 2016;2016:3136024. doi:10.1155/2016/3136024
29. Gu Y, Li J, Guo D, et al. Identification of 13 key genes correlated with progression and prognosis in hepatocellular carcinoma by weighted gene co-expression network analysis. Front Genet. 2020;11:153. doi:10.3389/fgene.2020.00153
30. Li Z, Wang Q, Peng S, et al. The metastatic promoter DEPDC1B induces epithelial-mesenchymal transition and promotes prostate cancer cell proliferation via Rac1-PAK1 signaling. Clin Transl Med. 2020;10(6):e191. doi:10.1002/ctm2.191
31. Xu Y, Sun W, Zheng B, et al. DEPDC1B knockdown inhibits the development of malignant melanoma through suppressing cell proliferation and inducing cell apoptosis. Exp Cell Res. 2019;379(1):48–54. doi:10.1016/j.yexcr.2019.03.021
32. Yang Y, Liu L, Cai J, et al. DEPDC1B enhances migration and invasion of non-small cell lung cancer cells via activating Wnt/β-catenin signaling. Biochem Biophys Res Commun. 2014;450(1):899–905. doi:10.1016/j.bbrc.2014.06.076
33. Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36(11):1461–1473. doi:10.1038/onc.2016.304
34. Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: wnt signaling and stem cell control. Science. 2014;346(6205):1248012. doi:10.1126/science.1248012
35. Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48(6):607–616. doi:10.1038/ng.3564
36. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013
37. Stankovic B, Bjørhovde HA, Skarshaug R, et al. Immune cell composition in human non-small cell lung cancer. Front Immunol. 2018;9:3101. doi:10.3389/fimmu.2018.03101
38. Anichini A, Perotti VE, Sgambelluri F, Mortarini R. Immune escape mechanisms in non small cell lung cancer. Cancers. 2020;12(12):3605. doi:10.3390/cancers12123605
39. Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi:10.1016/j.immuni.2013.10.003
40. Jin C, Lagoudas GK, Zhao C, et al. Commensal microbiota promote lung cancer development via γδ T cells. Cell. 2019;176(5):998–1013.e16. doi:10.1016/j.cell.2018.12.040
41. Story JY, Zoine JT, Burnham RE, et al. Bortezomib enhances cytotoxicity of ex vivo-expanded gamma delta T cells against acute myeloid leukemia and T-cell acute lymphoblastic leukemia. Cytotherapy. 2021;23(1):12–24. doi:10.1016/j.jcyt.2020.09.010
42. Faustino LD, Griffith JW, Rahimi RA, et al. Interleukin-33 activates regulatory T cells to suppress innate γδ T cell responses in the lung. Nat Immunol. 2020;21(11):1371–1383. doi:10.1038/s41590-020-0785-3
43. Theoharides TC. Mast cells and pancreatic cancer. N Engl J Med. 2008;358(17):1860–1861. doi:10.1056/NEJMcibr0801519
44. Nielsen HJ, Hansen U, Christensen IJ, Reimert CM, Brünner N, Moesgaard F. Independent prognostic value of eosinophil and mast cell infiltration in colorectal cancer tissue. J Pathol. 1999;189(4):487–495. doi:10.1002/(sici)1096-9896(199912)189:4<487::Aid-path484>3.0.Co;2-i
45. Davis BP, Rothenberg ME. Eosinophils and cancer. Cancer Immunol Res. 2014;2(1):1–8. doi:10.1158/2326-6066
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.