Back to Archived Journals » Vaccine: Development and Therapy » Volume 5
Hendra and Nipah viruses: pathogenesis, animal models and recent breakthroughs in vaccination
Authors Weingartl H
Received 12 April 2015
Accepted for publication 13 June 2015
Published 28 September 2015 Volume 2015:5 Pages 59—74
DOI https://doi.org/10.2147/VDT.S86482
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Don Diamond
Hana M Weingartl
National Centre for Foreign Animal Disease, Canadian Food Inspection Agency, Winnipeg, MB, Canada
Abstract: Hendra and Nipah viruses are two highly pathogenic zoonotic members of the genus Henipavirus, family Paramyxoviridae, requiring work under biosafety level 4 conditions due to a lack of effective therapy and human vaccines. Several vaccine candidates were protective in animal models: recombinant vaccinia virus expressing Nipah virus (NiV) F and G proteins in hamsters against NiV; recombinant ALVAC–NiV F and G in swine against NiV; recombinant Hendra virus (HeV) soluble G protein (sGHeV) against HeV and NiV in cats, ferrets, horses, and African green monkeys (AGM); recombinant vesicular stomatitis virus-based vectors expressing NiV F or G against NiV in hamsters and ferrets; measles virus-based NiV G vaccine candidate in hamsters and AGMs against NiV; and adenoassociated virus expressing NiG protein, which protected hamsters against NiV. The sGHeV was licensed for use in horses (Equivac HeV®) in 2012. It is the first vaccine candidate licensed against a biosafety level 4 agent. With the development of suitable animal models (ferret, hamster and, importantly, AGM), progress can be made toward development of a human vaccine.
Keywords: henipavirus, equine, swine, human infection, animal models, vaccine candidates
Introduction
Hendra and Nipah viruses: reservoir, transmission, epidemiology
Henipaviruses are considered to be bat viruses, found to be associated with bats from the families Pteropididae, Phystolomidae, and Mormoopidae. Pteropus bats serve as a reservoir for two zoonotic viruses: Hendra virus (HeV) and Nipah virus (NiV), classified into genus Henipavirus in the family Paramyxoviridae, together with the recently discovered nonpathogenic Cedar virus.1,2 Bats infected with henipaviruses do not develop an apparent disease, as confirmed by experimental infections with HeV in pregnant and nonpregnant Pteropus poliocephalus and P. alecto,3–5 and NiV in P. poliocephalus.6 Experimentally or naturally infected bats, however, seroconvert and shed the virus in urine, saliva, and uterine fluid.6–10 There is a greater than 40% prevalence of antibodies to HeV in Australian pteropid bats11,12 with viral RNA and infectious virus detected in urine samples collected under the roosting flying foxes,10 and in free-living colonies of P. alecto (fetus) and P. poliocephalus (fetus and uterine fluid).7 NiV antibodies were detected in Cynopterus brachyotis, Eonyveteris spelaea, Scotophuilus, P. vampyrus, P. hypomelanus, P. giganteus, and P. lylei; the reservoir status of the bats was confirmed by the detection of viral RNA or by virus isolation.8,13–15
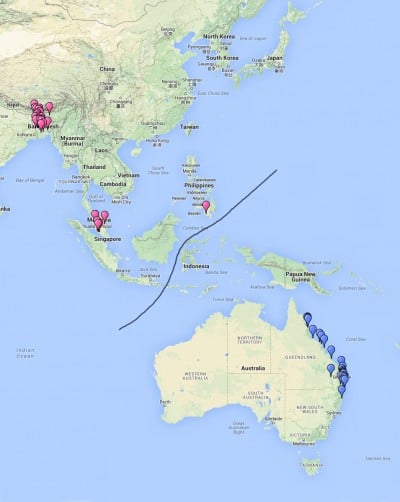
The geographical locations of the up to date HeV and NiV outbreaks correspond to the geographical distribution of the natural reservoir species in Australia (Hendra) and Southeast Asia (Nipah), respectively (Figure 1).
HeV and NiV are able to successfully cross interspecies barriers and infect a wide range of host species; in this respect, they are unusual among paramyxoviruses. Spillover of HeV and NiV from their natural reservoir to humans, pigs, or horses with secondary transmission to humans is sporadic, and appears to be seasonal, possibly related to stress in bats due to their natural life cycle, such as mating and breeding, enhanced by environmental pressures due to a loss of habitat.
Henipaviruses were discovered in 1994 in Australia during an outbreak of severe respiratory disease in horses, with 14 animal deaths. During the outbreak, virus transmission also occurred to a horse trainer and a stable hand providing care for the sick horses.16–18 The etiological agent was initially named equine morbillivirus, and it was later renamed (and reclassified) after the geographical location of the first isolate to HeV. None of the subsequent sporadic outbreaks in horses was as extensive as the first recognized one.19 A total of 90 deaths in horses was recorded over the next 20 years (between 1994 and 2014), with most of the self-limiting outbreaks occurring between May/June and September/October.10,20 Horses are suspected to become infected by grazing on pastures contaminated by bat urine, food debris (masticated fruit), or birthing material. Horse-to-horse transmission occurs via respiratory secretions – either due to direct contact among horses, or as a fomite. The virus transmission is not considered very effective unless a superspreader is involved, as was the case in the initial outbreak, or in the 2008 outbreak at the veterinary clinic.21,22 Similarly, transmission from horses to humans is not very efficient, since it requires a high degree of exposure to respiratory secretions (possible open abrasions or cuts on the skin, or direct inoculation of the mucosa – eg, in the eye).10,21 From the seven reported clinical cases of human HeV infections, four were fatal. All cases involved either veterinarians or people caring for very ill or moribund horses, or those performing necropsies of dead animals. However, the last two human cases were initially due to treating a horse that did not show clinical signs of Hendra disease at that time, but it was infectious to another horse prior to developing clinical signs.22,23 This corresponds with experimental findings of presymptomatic shedding in horses infected with HeV.24 Human-to-human transmission of HeV was not reported, and only one asymptomatic infection of a dog was reported in connection with one of the horse cases.10
NiV was first isolated from the cerebrospinal fluid of a patient from the village of Nipah close to Ipoh in Malaysia during an outbreak of viral encephalitis in 1998/1999.25 The route of NiV transmission in this, and an associated outbreak in Singapore, was initially from bats to pigs via pig feed contaminated by bat urine or saliva.8,9,26,27 Transmission from pigs to humans, confirmed by sequence analysis of human and swine isolates,28 occurred by direct exposure to the contaminated body fluids or tissues of pigs,12,29 or it was airborne by large droplets, as supported by the detection of NiV in the upper and lower respiratory tract of the infected pigs.26,30,31 There were a total of 265 human cases with 105 deaths reported.25,27,32–34 The outbreak was controlled by culling in an excess of one million pigs, resulting in substantial economic loss.35
NiV was first identified as an etiological agent of human encephalitis and severe pulmonary disease in the early outbreaks of 2001 in Bangladesh, and retrospectively also in India’s West Bengal. The virus causes seasonal, relatively small outbreaks with mortality rates as high as 75% compared to the 40% seen in the outbreak in Malaysia, with an estimated 400 cases between 2001 and April 2013 in this area of the Indian subcontinent.36–40 Outbreaks in West Bengal and Bangladesh start in December and cease by April, overlapping with the palm sap harvest and the breeding season of Pteropus spp. bats. Stress associated with breeding leads to increased shedding of NiV in bats, resulting in a high risk for spillover of the virus.14,21,41–43 Consummation of fruit or, importantly, of the date palm sap contaminated with bat saliva and/or urine is the primary risk factor for human NiV infection in Bangladesh and India.13,15,44,45 There is epidemiological evidence that in a few cases in Bangladesh, NiV may have been transmitted from infected domestic animals (cow, goat, pig) to humans.13,21 One important characteristic of the outbreaks in India and Bangladesh caused by the NiV-B lineage of the virus is person-to-person transmission.13,36,38,39 In cases where human-to-human transmission was documented, the clinical disease developed 6–11 days postexposure.46,47 The transmission requires very close contact with an ill or deceased person, usually by family members or attending medical staff, and larger clusters were found to be associated with a superspreader.39 NiV RNA was detected by reverse transcription polymerase chain reaction in the urine of several patients.36
In 2014, an outbreak of a henipavirus (suspected to be NiV based on serology and partial genome sequence) occurred in the Philippines with horse-to-human and human-to-human transmission, resulting in 17 human cases with 82% case-fatality, the death of ten horses, and possibly one dog and several cats, which were fed the horse meat. It is suspected that similarly to infections with HeV in Australia, the source of horse infections were bats from the family Pteropodidae roosting near one of the villages. The outbreak started by the sudden death of several horses due to a neurological disease, and the first human cases were infected during the slaughter and consumption of undercooked horse meat. Several human cases (primarily involving the medical staff), however, acquired the infection through human contact.48 There is field evidence that horses, cats, and dogs were infected with NiV also during the 1998/1999 outbreak in Malaysia.26,49
HeV and NiV: classification, virus proteins, and virus–host interaction
HeV and NiV virus belong to the genus Henipavirus, subfamily Paramyxovirinae, family Paramyxoviridae (enveloped, nonsegmented, negative-strand RNA viruses). Their genome organization resembles viruses found in the Respirovirus and Morbillivirus genera.50–52 A prominent molecular feature of henipaviruses is their large genome when compared to other paramyxoviruses: 18,234 nucleotides for HeV and 18,246 nucleotides for NiV.53 Nucleotide variation and the associated amino acid variation range detected between different NiV isolates, along with some differences in clinical factors, led to the classification of NiV into two genotypes: Malaysia (NiV-M) and Bangladesh (NiV-B).54–58
The genomic RNA of henipaviruses is encapsidated by the nucleoprotein (N), with the phosphoprotein (P), and RNA-dependent RNA polymerase (large protein; L) being minority proteins of the nucleocapsid as well.59 Figure 2 illustrates the structural arrangement of the NiV nucleocapsid in the virion. Infected hosts generally produce good non-neutralizing antibody titers against the N protein, making detection of anti-N antibodies suitable for enzyme-linked immunosorbent assay or Luminex assay, and of interest for the differentiating infected from vaccinated animals (DIVA) strategy in veterinary vaccine development.60,61 Location of the N gene at the very 3′ end of the genome makes it a sensitive target for RNA detection in infected cells due to the abundance of the transcripts from it.62
The nucleocapsid interacts with matrix (M) protein, located at the inner surface of the virion’s lipid envelope. The M protein, which drives the virus budding, also interacts with the cytoplasmic tale of the fusion (F) protein, thus stabilizing the virion.51,63,64 The F protein (type I membrane protein) and the attachment G glycoprotein (type II membrane protein) embedded in the lipid bilayer of the envelope form projections on the virion surface and elicit the development of neutralizing antibodies65,66 in all infected hosts, including bats. That makes the F and G proteins primary candidates as immunizing antigens in vaccine development.67
Henipavirus fusion is a pH-independent process, but it requires proteolytic cleavage of the F protein (F0) into two subunits (F1 and F2). The F0 precursor expressed on the cell surface is endocytosed in clathrin-coated vesicles, cleaved by endosomal proteases known as cathepsins, and the functionally mature F protein is transported back to the plasma membrane,68 where it forms trimers.69 Direct plasma membrane fusion and macropinocytosis were identified as the henipavirus mode of cell entry,59,70,71 and this may be cell type specific. Plasma membrane fusion of infected cells with uninfected ones further facilitates virus spread throughout the host, and contributes to the pathology of henipavirus disease. Epithelial and endothelial syncytia, and multinucleated cells (mostly macrophages and dendritic cells), were observed in the tissues of humans and other host species.6,49,72–75
Henipavirus fusion by the F protein also requires, in addition to cleavage of the F protein, activation of this protein by the second viral glycoprotein, the G protein.76 Henipavirus G glycoproteins form covalently linked dimers noncovalently associated into tetramers.77–79 In contrast to attachment glycoproteins of other paramyxoviruses, henipavirus G proteins lack both hemagglutinin and neuraminidase activities.66,80 HeV and NiV glycoprotein G is the virus attachment protein recognizing ephrin B2 and ephrin B3 as cellular receptors,81,82 and it may also bind with lower affinity to a C-type lectin on endothelial cells in the lymph nodes and liver.78 Ephrin B2 is important in mammalian host development and it is a highly conserved protein across mammalian species, expressed on lymphocytes, neurons, smooth muscle cells, and endothelial cells surrounding small arteries, corresponding with virus distribution in an infected host as determined by immunohistochemistry.30,56,73,83–85 The cellular function of ephrin B2 is to regulate processes such as neurogenesis, angiogenesis, proliferation, and remodeling, as well as immune activation and bone formation.86–88 HeV G protein has a somewhat lower affinity for ephrin B3 compared to the NiV G protein.81 This may play a role in the somewhat different pathology in human infections, where NiV with its higher affinity for this receptor than HeV, causes fatal neurological disease with severe brain stem dysfunction,34,73 although the last human fatal case of HeV had extensive brain involvement.23 Interestingly, in experimentally infected swine, HeV invasion of the central nervous system (CNS) was limited in the early stages to the olfactory bulb alone,75 and with a lower inoculation dose, HeV did not reach the CNS (Pickering B, Weingartl HM, unpublished data, 2015). Using ubiquitous and conserved proteins as receptors, henipaviruses not only have a wide host range, but they also infect a wide range of tissues and organs within each individual host species.
The P gene of henipaviruses encodes three nonstructural proteins (C, V, and W) in addition to the phosphoprotein P. The P protein is critical for virus replication,89,90 and through its interaction with cellular proteins, it also modulates cell signaling. Some of the functions encoded in the N terminus of the protein are shared with the V and W proteins due to an identical N-terminal portion of the three proteins. The V protein is produced by the cotranscriptional addition of one nontemplated G at the editing site, and the W protein by insertion of two nontemplated Gs.91–94 The proteins are located in the cytoplasm, except for the W protein being detected also in the nucleus in some cell types. The C protein is encoded by a separate internal open reading frame within the P gene,91,92,95 and it localizes predominantly into the perinuclear region.94,96 The P, W, and V proteins hinder the interferon JAK–STAT signaling pathway by binding to STAT1 and preventing its translocation into the nucleus.96–98 Nuclear localization of NiV W impairs the TLR3/TRIF pathway by blocking TRIF-mediated activation of interferon regulatory factor (IRF)-3 responsive promoter, ultimately interfering with the induction of interferon (IFN)-β and other molecules controlled by this pathway.99 NiV V proteins bind to the MDA5 helicase along with LGP2 to suppress RIG-I-like (RLR) signaling, thereby inhibiting the downstream signaling events also leading to IFN-β synthesis.100,101 NiV C exhibits inhibitory activity against TRL7/9-dependent IFN-α induction by binding to IKKs and inhibiting phosphorylation of IRF-7,102 as well as influencing IFN-β and antiviral gene expression.103 A study by Mathieu et al104 suggested that the C protein can regulate cytokine balance in transfected cells. The nonstructural proteins are involved in the NiV life cycle by regulating replication and evading the innate immune response,105,106 as confirmed by in vivo reverse genetics studies in animal models.107,108
The evasion of the IFN system appears to be cell specific, since NiV will induce IFN type I and other innate cytokines in endothelial cells, an important in vivo target for henipaviruses,93,104 and there is the possibility that cells may employ alternative pathways to establish an antiviral state.109 The molecular aspects of henipavirus replication and its interaction with the host cell are – with some differences between HeV and NiV, and between individual nonreservoir host species – reflected in the clinical disease, pathogenesis, pathology, and immune response.
Infections in natural, nonreservoir hosts with henipaviruses
Human henipavirus infections
With only few human HeV disease cases, the clinical symptoms and pathology of henipavirus infections in humans are summarized mainly based on clinical infections with NiV, since there appear to be common general characteristics and similarities between Hendra and Nipah infections, as well as between NiV infections with the Malaysia and Bangladesh genotype. Symptomatic infections present mainly as severe acute encephalitis, with severe pulmonary involvement often being reported from Bangladesh.23,32,34,47,110,111 Infections with a virus causing the more severe respiratory form may be potentially associated with increased shedding and the observed human-to-human transmission with the NiV-B genotype.13,21,36 Disease onset is characterized by fever and headache, followed by varying degrees of altered consciousness in the majority of cases. Some patients suffer from, nausea, vomiting, muscle pain, and involuntary muscle movements. In fatal cases, death likely due to severe brain stem involvement generally occurred within 1 week or 2 weeks after the onset of symptoms.13,33,34,36,47 The neurological signs are supported by pathological findings: brain damage due to the infection of small blood vessels accompanied by vasculitis with thrombosis; hemorrhage and frequent adjacent necrosis; as well as direct infection of specific groups of neurons.73,112 Henipaviruses can persist in the CNS of some of the infected individuals causing neurological relapse.113,114 In late-onset encephalitis, the target cells are neuronal only, with no vascular involvement.112,114
Although there is only a low level of viremia detected in humans, viremic spread of NiV appears to be the main route of CNS invasion in humans, and the virus can be isolated from the cerebrospinal fluid.25,73 NiV replicates to a low level in human dendritic cells, and it can spread mechanically through attachment to monocytes and lymphocytes115 and putatively via cell-free viremia.73 Infection of the endothelial cells of the small blood vessels is a hallmark of henipavirus infections, and it contributes to virus spread into the parenchyma of several organs – importantly, also through the blood–brain barrier and the blood–air barrier contributing to virus spread and pathology in the brain and lungs.72,116 Epithelial cells can also be infected with henipavirus, interestingly either from the basal membrane or by fusion with the neighboring epithelial cells, supporting a hypothesis for viremic spread to the lungs besides via the airway route.64
People infected with henipaviruses develop antibodies against the virus that may last for years.13,18,19,36,113,117 Based on serological evidence, subclinical infections were reported in Malaysia and Singapore.118
Henipaviruses in horses
NiV is suspected to infect horses in the 2014 outbreak in the Philippines; however, the etiological agent was identified retroactively through human samples, and thus no pathological examination or sample collection took place.48 Only partial sampling (brain, spinal cord) was conducted for one horse in the Malaysian outbreak with immunohistological evidence of virus infection.49
HeV-infected horses develop severe, acute, febrile respiratory disease sometimes accompanied by facial swelling, ataxia and, terminally, copious frothy nasal discharge, increased rectal temperature (41°C), and profuse sweating, which can resemble African horse sickness, for example. Neurological signs with clinical features such as hypersensitivity, ataxia, disorientation, head pressing, and stranguria predominated in more recent outbreaks.16,17–19,22 Shedding of the virus in oral secretions in HeV-infected horses prior to the onset of clinical signs began to be suspected during the 2008 outbreak, and it was confirmed experimentally starting 48 hours postinfection. After the onset of a clinical disease, the virus can also be detected in urine and feces.3,10,22,24
Similar to human cases, predominant lesions in the HeV infection of horses include vasculitis with syncytial cells in the vascular endothelium, fibrinoid degeneration, and necrosis, most markedly in the lymphatic vessels and small blood vessels in major organs such as the lungs, brain (including meninges), spleen, lymph nodes, and kidney, but which are also detectable in the nasal mucosa, liver, heart, stomach, intestine, uterus, and ovaries. Pulmonary lesions in horses are particularly extensive, with severe necrotizing alveolitis with marked fibrinous alveolar exudates, corresponding with severe respiratory clinical signs in infected animals. Syncytial cells were observed in addition to endothelial cells in respiratory epithelial and lymphoid cells as well. Viral antigen was detected in the affected organs and tissues, including the brain and choroid plexus.24,119,120
While some HeV-infected animals remain asymptomatic, horses that have survived an acute HeV infection and that have seroconverted may have moderate focal nonsuppurative meningoencephalitis, with the HeV antigen and viral RNA still detectable several weeks after the resolution of clinical signs. Persistent infection of horses has not been identified.16,119,121
Henipaviruses in pigs
HeV can, under experimental conditions, infect swine by the oronasal route, causing a disease similar to NiV, although with limited invasion of the CNS;75 however, natural infections of pigs with HeV have not been reported.122
In contrast, NiV-infected pigs were the source of human infections in the first recognized outbreak in Malaysia in 1998/1999.28 The virus appears to be highly contagious in pigs with an infection rate close to 100% in affected farms, but with low mortality (1%–5%). Direct, and possibly also airborne, exposure to secretions from infected animals was one of the presumed modes of transmission of NiV among pigs, supported by the detection of NiV in the epithelium of the upper and lower respiratory tract, as well as in the lumen of the airways.26,30,31,49,84 Another important route of infection is oral by ingestion of contaminated material (especially from bat to pig), likely resulting in less severe clinical signs observed in the field. Generally, the disease was mild with nonspecific clinical signs which, in addition, varied depending on the age of the pig. However, if pigs developed severe disease, neurological and/or respiratory signs were the most frequently observed.26 In the experimentally infected animals, severe clinical signs were observed in subcutaneously, nasally, or oronasally inoculated pigs compared to orally inoculated ones. About 20%–40% of piglets developed CNS signs requiring euthanasia, either due to viral encephalitis or to secondary bacterial meningitis.30,31,119,123,124 Pathological observations, especially microscopical, correspond with human or horse NiV infections.30,49,84,119,125
Besides the ability of the swine host to mount an effective protective immune response in the majority of animals in a natural setting, there are two other characteristics that are different from human or horse infections with NiV: direct invasion of the brain via the cranial nerves;30 and transient immunosuppression early postinfection, linked to the ability of NiV to infect a range of porcine immune cells, such as dendritic cells, monocytes, macrophages, natural killer cells, and CD8+ T-cells.31,116 This, together with the ability of henipaviruses to evade the IFN system, may lead to delayed or modified immune cell signaling, in turn leading to a less efficient innate immune response and less efficient transition to the adaptive immune response. Nevertheless, pigs develop protective neutralizing antibodies against Nipah only with a short delay.31,119 A protective immune response in pigs requires both humoral and immune cell memory (Pickering B, Weingartl HM, unpublished data, 2015), in agreement with the pilot data from a previous vaccine efficacy study.124 Serum antibodies against NiV appear to be long lasting, as suggested by the postoutbreak investigation in Malaysia.
The current understanding of routes of infection and virus spread throughout the host infected with henipaviruses is briefly summarized in Figure 3. Despite some differences in the pathogenesis and pathology between species, there are common characteristics of henipavirus infections, such as the development of viremia; the involvement of the respiratory, central nervous, and immune systems; vasculitis; the formation of syncytia in the endothelium; the infection of dendritic cells, etc. Considering the routes of infection, mucosal immunity may be important early postinfection; however, this has not been widely explored, even in terms of vaccine design.
Animal models of henipavirus infection employed in vaccine development and efficacy studies
Approval of human vaccines requires that the vaccine candidate is found to be efficacious in at least two animal models representing a human disease, preferably including a nonhuman primate model, while for veterinary vaccines, the requirement is efficacy in the target species. However, vaccine efficacy testing for large animals under biosafety level 4 conditions is extremely difficult and constrained. Consequently, the two animal models rule was considered when licensing the HeV vaccine for horses (Equivac HeV®) in Australia, based on the outcomes of protection studies in ferrets against HeV, and cats, ferrets, and nonhuman primates against NiV. In addition to horses, guinea pigs and ferrets were used in parallel to complement the efficacy testing.126
Equine and swine target species models
The two large animal models used in vaccine efficacy testing are both based on studies looking into the susceptibility, pathogenesis, and transmission described in the previous section. Oronasal inoculations with 2×106 median tissue culture infective dose (TCID50)/animal of HeV in a horse model are fatal,3,16,17,24,119,120,126 in contrast to NiV infection in a pig model, where only 20%–40% of animals require euthanasia due to CNS signs. This putative fatality rate is already much higher than the one observed during the 1998/1999 outbreak in Malaysia, and a lethal model in this species may not be possible. However, all animals in the existing model are infected and have a high virus load in their tissues and oronasal secretions.30,31,119,124 NiV vaccine efficacy trials in young pigs (4 weeks of age at immunization) were conducted employing this model using intranasal (IN) inoculation with 2.5×105 plaque forming units (PFU) per piglet at the age of 10 weeks.124
Naturally infected species models
Cats can be naturally infected with NiV,48,49 which makes them an a priori suitable candidate as a model for vaccine efficacy studies. Cats have been shown to be highly susceptible to HeV or NiV infection under experimental conditions with clinical and pathological features similar to human disease: HeV-infected cats exhibit fever and changes in behavior, accompanied with respiratory disease often leading to death. All routes of inoculation – oral, nasal, and subcutaneous – lead to acute clinical disease and death using doses of 5×103 or 5×104 TCID50 per animal. In-contact animals were also infected and featured similar outcomes as the inoculated ones. HeV produced significant respiratory signs, including dyspnea with increased temperature, typically dropping 24 hours before death. Severe interstitial pneumonia, vasculitis (lung, gastrointestinal, and lymphoid systems), and syncytia in the endothelium were detected postmortem.3,127,128 Cats infected with NiV (5×102–5×104 TCID50 per animal) oronasally or subcutaneously had, on postmortem examination and in addition to lesions observed with HeV, necrosis of the lymphatic tissues, degeneration of neurons, and moderate meningitis.123,129 Experimental infection of a pregnant cat (unintentional) provided further evidence that the virus can be transmitted to the fetus.130
Since dogs can be naturally infected with henipaviruses, and although they are not considered a secondary reservoir,131 there is a possibility that ferrets (another canine species) may be a suitable model for henipavirus, which was eventually considered.132,133 Several protection or vaccine efficacy studies employing different vaccine platforms demonstrated that ferrets are a reliable challenge model for henipaviruses.126,134–136 It appears that a minimal NiV infectious dose to infect all inoculated ferrets oronasally is 5×103 TCID50 with highest virus load in the lungs and spleen, detected in all animals infected with this and one log10 higher dose. All four animals in this study by Bossart et al132 also had the virus in the olfactory section of the brain. NiV was detected mainly in the neurons, and detection of the virus antigen in ependymal epithelium and the arachnoid membrane of the meninges would indicate viremic spread of the virus into the CNS. The HeV model (oronasal inoculation with 5×103 TCID50) was described by Pallister and colleagues.133,134 Recently, a recombinant HeV-expressing GFP has been reported to facilitate further pathogenesis studies.137
Both models represent human henipavirus histopathology well, with typical multisystemic vasculitis including the lung, spleen and, to a lesser extent, the brain. The lung involvement also warranted the use of the HeV/ferret model for the equine vaccine efficacy trials as a control.126,134
Small animal models
Following the initial outbreak of HeV in Australia, an early experiment focusing on species susceptibility to HeV tested many animal models including mice, guinea pigs, rats, chickens, rabbits, cats, and dogs, with only guinea pigs and cats developing the disease,128 although it has to be noted that the dogs were vaccinated against canine distemper, a virus from the closely related genus Morbillivirus. Further studies, unfortunately, determined that the outcome of guinea pig infections with henipaviruses is not consistent, and that the guinea pig model is not reliable for vaccine efficacy studies of HeV vaccine candidates, and is even less reliable for efficacy studies of NiV vaccine candidates.4,6,74,138 Interestingly, work by Williamson et al4 showed that HeV was transmissible to the guinea pig fetus during gestation.
Mice, although originally found to be refractory to both HeV and NiV,74,128 are being recently developed as disease models, employing animals with an impaired immune response. For example, IFN-α receptor knockout, or aged C57BL/6 and BALB/c mice were reported to be susceptible to henipavirus infections.139,140 An intriguing mouse model to study human lung tissue pathogenesis in NiV infections has been developed using human lung xenograft in immunodeficient NSG mice.141 These models would not be considered suitable for vaccine efficacy studies due to impairments in their immune response. However, genetically defined mice are an excellent tool to elucidate specific aspects of henipavirus pathogenesis.140,142 Mice with functional immune responses were used in a number of proof of principle immunization studies for different vaccine candidate platforms.143–145
Golden (Syrian) hamster is the first and currently the only small animal model used for vaccine efficacy testing.146 The original model was developed to mimic human NiV encephalitis74 and, more recently, it was modified to simulate the respiratory involvement during henipavirus infections for both NiV and HeV,147 and to study transmission.148–150 The first pathogenesis study in hamsters with HeV indicated that the animals are highly susceptible to the virus: a dose of 103 PFU per animal was 100% fatal when inoculated intraperitoneally (IP). Inoculation with 100 PFU indicated that there may be age differences in susceptibility, with survivors in the older animals group.151 IN and IP inoculations of hamsters with higher doses both result in clinical signs and death, although delayed course of the disease relative to the IP inoculation was observed with the same dose of virus administered IN.74 IN inoculation with a high dose of NiV, and especially HeV, resulted in severe acute respiratory distress in the infected hamsters.147 High IP NiV dose or IN dose (range of 107–105 PFU/TCID50) resulted in acute encephalitis. In the high-dose IN inoculation, the virus accessed the brain via cranial nerves from the nasal cavity early postinoculation,152 similar to the neuroinvasion of NiV in pigs.30 The high-dose inoculated animals developed tremor and limb paralysis 24 hours before death, with virus detected in the urine, brain, spinal cord, lungs, kidney, spleen, liver, and heart. Low-dose (10 PFU and 102 PFU) IN inoculation was not uniformly fatal, and survivors developed high levels of neutralizing antibodies by 29 days postinoculation (dpi).74 The IN inoculation with a low dose of HeV or NiV resulted in systemic spread and invasion of the brain by the virus crossing the blood–brain barrier.147 There may be some differences in the pathogenesis between the Malaysian and Bangladesh genotypes of NiV, as different lesion severities were observed in IP-inoculated animals between the two, but not in the IN-infected animals.57,58
Nonhuman primate models
Squirrel monkeys were found to be only partially susceptible to NiV, with most challenged animals not developing clinical signs – even with a high challenge dose – and viral RNA was detectable in only some organs of some animals.153
The African green monkey (AGM) model well resembles disease and pathology in humans, and the animals are susceptible to both HeV (dose in a range of 105 PFU per animal) and NiV (dose range of 104 PFU) infection via the intratracheal route.85,154–156 NiV infection in this model was well analyzed, indicating pulmonary and CNS involvement, with microscopic brain lesions and antigen distribution in correlation with findings in humans. The model does not appear to be 100% lethal, even with the IP dose of 105 TCID50, but all inoculated animals are infected with detectable virus in swabs, urine, and multiple organs, and they develop viremia.154,156 The oronasal route of inoculation with 106 or 108 TCID50 was also not uniformly lethal.157 The amount of viral RNA detected in peripheral blood mononuclear cells (PBMCs) is rather significant (106–107 copy numbers/mL),156 and it may indicate viral replication in some subpopulations of AGM leukocytes. Lymphocyte depletion in the spleen, lymph nodes, and tonsils was observed in IP-inoculated AGMs.157 Interestingly Geisbert et al154 found more virus associated with the red blood cell fraction than in PBMCs and granulocytes. A somewhat higher dose of HeV per animal (4×105 TCID50) resulted in uniform lethality in the study involving 12 AGMs. The animals developed disease and pathology consistent with human disease.85 However, vaccine efficacy studies indicated that the HeV-infected AGM model is similar to NiV, not a lethal challenge model.155 Nevertheless, the availability of the AGM model is crucial for the development of human vaccines, and additional correlates of infection can be developed for protection studies – eg, a lack of azotemia.156
Vaccine candidates
Reports on vaccine candidates tested in animals for their efficacy against NiV or HeV challenge are summarized in Table 1. As many of them can be considered for both human and veterinary vaccines, there is no distinction made in Table 1 based on their intended use.
An important strategy, when considering the protection of human population against zoonotic viruses, is interruption of the transmission cycle at the level of the intermediate host, or natural reservoir, if practical. Abolition or a very significant reduction of shedding following the challenge of vaccinated animals is considered a critical requirement for the efficacy of veterinary vaccines. The advantage of the vaccination of livestock or pets is that the vaccination also protects them. The second advantage of this approach is that the licensing of veterinary vaccines can generally take less time than the licensing of human vaccines. Licensing of the Hendra vaccine for horses (Equivac HeV®; Zoetis, Inc., Florham Park, NJ, USA) in Australia in November of 2012 is the best example. Following the fatal human case of HeV infection, Australian health authorities decided in 2009 to pursue rapid licensing of an equine vaccine for HeV. It took a total of 2 years from the decision to licensing. The successful candidate, soluble HeV G protein (sGHeV), which was first tested in cats in 2006,129 was shown to be protective against HeV in ferrets134 prior to testing in horses.126 Following the successful protection experiments in AGMs against both HeV and NiV,155,158 sGHeV may be potentially considered as a vaccine candidate for humans in the future. Even though the AGM model is not 100% lethal, additional correlates of protection and infection can be employed to assess protection,154–156,158 as is currently being considered for swine NiV efficacy tests, where a lack of viral RNA presence in tissues – and, importantly, a lack of shedding postchallenge – as well as cellular and humoral immune response are considered (Pickering B, Weingartl HM, unpublished data, 2015).124 Very interestingly, vaccination with sGHeV protein was found to be protective against NiV in number of species,129,135,158,159 but adenoassociated virus vector expressing the NiV G protein yielded only partial protection against HeV challenge, although it was fully protective against the NiV challenge.160 An adjuvant suitable for the target species has to be part of the vaccine formulation for the sGHeV-based vaccine candidates to offer full protection: shedding of viral RNA was detected in cats,159 but not in other tested species vaccinated with sGHeV and challenged with NiV.
Several recombinant virus vaccine candidates were also developed, some of them with no or limited replication in the target host, such as ALVAC, adenoassociated virus, or VSV-delta G requiring GInd for replication.124,136,160 The very first vaccine candidate tested was live recombinant vaccinia virus expressing NiV F or G protein. Besides demonstrating full protection, this study also indicated that neutralizing antibodies are protective in hamsters after passive transfer.146 The canarypox-based ALVAC candidate was fully protective with sterile immunity in the swine as a target species when both F and G antigen-expressing ALVAC preparations were used for immunization, as no viral RNA was detected in swabs or tissues.124 Vaccination employed a primary and boost immunization regimen for vaccinia, as well as for the canarypox-based vaccine candidate. The next set of recombinant virus candidates based on VSV attempted to eliminate the need for boost, and when coding for the NiV F or G protein, those candidates were protective against NiV challenge,136,161,162 importantly also in AGMs during a pilot vaccine efficacy study.163 Immunization with rVSV protein expressing NiV N protein generating only non-neutralizing antibodies was partially protective in the hamster model, perhaps suggesting some role for non-neutralizing antibodies as well. More likely though, this indicated that the cellular immune response plays a role in protection.161 One of the recent vaccine candidates based on the measles virus platform is protective both in hamsters and in AGMs, and it is likely intended for human use.157
Conclusion
Vaccine efficacy studies clearly indicate that the prevention of henipavirus disease by vaccination would be the best measure, considering that although numerous antivirals have been tested up to date in vitro, the in vivo efficacy was not very encouraging. Ribavirin and chloroquine were tested experimentally in animals, and they were also used for the clinical emergency treatment of humans; however, there was no conclusive benefit.23,85,110,133,164,165 The ability of henipaviruses to inhibit IFN response at the level of induction and signaling may, to a certain extent, explain the lack of effectiveness of ribavirin treatment.
Neutralizing antibodies have long been recognized as protective,135,146,151 and they are considered for postexposure treatment. Administration of the monoclonal antibody m102.4, which targets an epitope in the G protein receptor binding site166 and is thus effective against both HeV and NiV, provided full protection.132,167 Timing of the administration and the amount of antibody required for full protection, unfortunately, limit the use of this approach.
There are essentially no studies focusing on adaptive immune responses against henipaviruses. Our limited understanding comes from vaccination/immunization studies and they have focused on correlates of protection. For quite some time, the protection offered by vaccination was considered to be solely due to the presence of high levels of neutralizing antibodies prior to the challenge.146 Indications that adaptive cellular response may be important came from a study employing live recombinant vaccine ALVAC in swine. Vaccinated pigs had good IFN-γ response in vaccinated PBMCs, indicative of immune cell memory.124 Later, a study in hamsters using immunization with the rVSV-NiN protein showed partial protection despite a lack of neutralizing antibodies, again supporting the role of the cellular immune response in protection.161 In addition, protection against HeV was demonstrated in horses with low neutralizing antibody titers of only 16/32.126 Overall, it appears that depending on the host species, elicitation of both humoral and cell immunity memory may be required for vaccine efficacy.
Vaccination is, without a doubt, the best option for the protection of humans and animals against henipaviruses. With the Equivac HeV® vaccine licensed for use in horses in Australia in 2012, HeV became the first biosafety level 4 agent against which a vaccine was made available.
Acknowledgment
The author would like to thank Dr B Pickering for inputs into the Reference section and Figure 1.
Disclosure
The contents and opinions expressed in this review are solely the responsibility of the author. The author reports no conflicts of interest in this work.
References
Drexler JF, Corman VM, Müller MA, et al. Bats host major mammalian paramyxoviruses. Nat Commun. 2012;3:796. | |
Marsh GA, de Jong C, Barr JA, et al. Cedar virus: a novel Henipavirus isolated from Australian bats. PLoS Pathog. 2012;8(8):e1002836. | |
Williamson MM, Hooper PT, Selleck PW, et al. Transmission studies of Hendra virus (equine morbillivirus) in fruit bats, horses and cats. Aust Vet J. 1998;76(12):813–818. | |
Williamson MM, Hooper PT, Selleck PW, Westbury HA, Slocombe RF. Experimental hendra virus infection in pregnant guinea-pigs and fruit Bats (Pteropus poliocephalus). J Comp Pathol. 2000; 122(2–3):201–207. | |
Halpin K, Hyatt AD, Fogarty R, et al; Henipavirus Ecology Research Group. Pteropid bats are confirmed as the reservoir hosts of henipaviruses: a comprehensive experimental study of virus transmission. Am J Trop Med Hyg. 2011;85(5):946–951. | |
Middleton DJ, Morrissy CJ, van der Heide BM, et al. Experimental Nipah virus infection in pteropid bats (Pteropus poliocephalus). J Comp Pathol. 2007;136(4):266–272. | |
Halpin K, Young PL, Field HE, Mackenzie JS. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J Gen Virol. 2000;81(Pt 8):1927–1932. | |
Yob JM, Field H, Rashdi AM, et al. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg Infect Dis. 2001;7(3):439–441. | |
Chua KB, Koh CL, Hooi PS, et al. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect. 2002;4(2):145–151. | |
Field H, Crameri G, Kung NY, Wang LF. Ecological aspects of Hendra virus. Curr Top Microbiol Immunol. 2012;359:11–23. | |
Young PL, Halpin K, Selleck PW, et al. Serologic evidence for the presence in Pteropus bats of a paramyxovirus related to equine morbillivirus. Emerg Infect Dis. 1996;2(3):239–240. | |
Field H, Young P, Yob JM, Mills J, Hall L, Mackenzie J. The natural history of Hendra and Nipah viruses. Microbes Infect. 2001;3(4):307–314. | |
Hsu VP, Hossain MJ, Parashar UD, et al. Nipah virus encephalitis reemergence, Bangladesh. Emerg Infect Dis. 2004;10(12):2082–2087. | |
Wacharapluesadee S, Boongird K, Wanghongsa S, et al. A longitudinal study of the prevalence of Nipah virus in Pteropus lylei bats in Thailand: evidence for seasonal preference in disease transmission. Vector Borne Zoonotic Dis. 2010;10(2):183–190. | |
Epstein JH, Prakash V, Smith CS, et al. Henipavirus infection in fruit bats (Pteropus giganteus), India. Emerg Infect Dis. 2008;14(8):1309–1311. | |
Murray K, Selleck P, Hooper P, et al. A morbillivirus that caused fatal disease in horses and humans. Science. 1995;268(5207):94–97. | |
Murray K, Rogers R, Selvey L, et al. A novel morbillivirus pneumonia of horses and its transmission to humans. Emerg Infect Dis. 1995; 1(1):31–33. | |
Selvey LA, Wells RM, McCormack JG, et al. Infection of humans and horses by a newly described morbillivirus. Med J Aust. 1995; 162(12):642–645. | |
Hanna JN, McBride WJ, Brookes DL, et al. Hendra virus infection in a veterinarian. Med J Aust. 2006;185(10):562–564. | |
Mendez DH, Kelly J, Buttner P, Nowak M, Speare R. Management of the slowly emerging zoonosis, Hendra virus, by private veterinarians in Queensland, Australia: a qualitative study. BMC Vet Res. 2014;10:215. | |
Luby SP, Gurley ES. Epidemiology of henipavirus disease in humans. Curr Top Microbiol Immunol. 2012;359:25–40. | |
Field H, Schaaf K, Kung N, et al. Hendra virus outbreak with novel clinical features, Australia. Emerg Infect Dis. 2010;16(2):338–340. | |
Playford EG, McCall B, Smith G, et al. Human Hendra virus encephalitis associated with equine outbreak, Australia, 2008. Emerg Infect Dis. 2010;16(2):219–223. | |
Marsh GA, Haining J, Hancock TJ, et al. Experimental infection of horses with Hendra virus/Australia/horse/2008/Redlands. Emerg Infect Dis. 2011;17(12):2232–2238. | |
Chua KB, Goh KJ, Wong KT, et al. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet. 1999;354(9186):1257–1259. | |
Mohd Nor MN, Gan CH, Ong BL. Nipah virus infection of pigs in peninsular Malaysia. Rev Sci Tech. 2000;19(1):160–165. | |
Chua KB. Nipah virus outbreak in Malaysia. J Clin Virol. 2003; 26(3):265–275. | |
AbuBakar S, Chang LY, Ali AR, Sharifah SH, Yusoff K, Zamrod Z. Isolation and molecular identification of Nipah virus from pigs. Emerg Infect Dis. 2004;10(12):2228–2230. | |
Chua KB, Bellini WJ, Rota PA, et al. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288(5470):1432–1435. | |
Weingartl H, Czub S, Copps J, et al. Invasion of the central nervous system in a porcine host by nipah virus. J Virol. 2005;79(12):7528–7534. | |
Berhane Y, Weingartl HM, Lopez J, et al. Bacterial infections in pigs experimentally infected with Nipah virus. Transbound Emerg Dis. 2008;55(3–4):165–174. | |
Lee KE, Umapathi T, Tan CB, et al. The neurological manifestations of Nipah virus encephalitis, a novel paramyxovirus. Ann Neurol. 1999; 46(3):428–432. | |
Paton NI, Leo YS, Zaki SR, et al. Outbreak of Nipah-virus infection among abattoir workers in Singapore. Lancet. 1999;354(9186):1253–1256. | |
Goh KJ, Tan CT, Chew NK, et al. Clinical features of Nipah virus encephalitis among pig farmers in Malaysia. N Engl J Med. 2000; 342(17):1229–1235. | |
Lam SK, Chua KB. Nipah virus encephalitis outbreak in Malaysia. Clin Infect Dis. 2002;34 Suppl 2:S48–S51. | |
Chadha MS, Comer JA, Lowe L, et al. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg Infect Dis. 2006;12(2):235–240. | |
Harit AK, Ichhpujani RL, Gupta S, et al. Nipah/Hendra virus outbreak in Siliguri, West Bengal, India in 2001. Indian J Med Res. 2006; 123(4):553–560. | |
Gurley ES, Montgomery JM, Hossain MJ, et al. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg Infect Dis. 2007;13(7):1031–1037. | |
Luby SP, Hossain MJ, Gurley ES, et al. Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001–2007. Emerg Infect Dis. 2009;15(8):1229–1235. | |
Homaira N, Rahman M, Hossain MJ, et al. Nipah virus outbreak with person-to-person transmission in a district of Bangladesh, 2007. Epidemiol Infect. 2010;138(11):1630–1636. | |
Reeder DM, Kosteczko NS, Kunz TH, Widmaier EP. The hormonal and behavioral response to group formation, seasonal changes, and restraint stress in the highly social Malayan Flying Fox (Pteropus vampyrus) and the less social Little Golden-mantled Flying Fox (Pteropus pumilus) (Chiroptera: Pteropodidae). Horm Behav. 2006;49(4):484–500. | |
Rahman MA, Hossain MJ, Sultana S, et al. Date palm sap linked to Nipah virus outbreak in Bangladesh, 2008. Vector Borne Zoonotic Dis. 2012;12(1):65–72. | |
Rahman SA, Hassan L, Epstein JH, et al; Henipavirus Ecology Research Group. Risk Factors for Nipah virus infection among pteropid bats, Peninsular Malaysia. Emerg Infect Dis. 2013;19(1):51–60. | |
Luby SP, Rahman M, Hossain MJ, et al. Foodborne transmission of Nipah virus, Bangladesh. Emerg Infect Dis. 2006;12(12):1888–1894. | |
Sazzad HM, Luby SP, Ströher U, et al. Exposure-based screening for Nipah virus encephalitis, Bangladesh. Emerg Infect Dis. 2015; 21(2):349–351. | |
Tan CT, Tan KS. Nosocomial transmissibility of Nipah virus. J Infect Dis. 2001;184(10):1367. | |
Hossain MJ, Gurley ES, Montgomery JM, et al. Clinical presentation of nipah virus infection in Bangladesh. Clin Infect Dis. 2008;46(7):977–984. | |
Ching PK, de los Reyes VC, Sucaldito MN, et al. Outbreak of henipavirus infection, Philippines, 2014. Emerg Infect Dis. 2015;21(2):328–331. | |
Hooper P, Zaki S, Daniels P, Middleton D. Comparative pathology of the diseases caused by Hendra and Nipah viruses. Microbes Infect. 2001;3(4):315–322. | |
Wang LF, Yu M, Hansson E, et al. The exceptionally large genome of Hendra virus: support for creation of a new genus within the family Paramyxoviridae. J Virol. 2000;74(21):9972–9979. | |
Wang YE, Park A, Lake M, et al. Ubiquitin-regulated nuclear-cytoplasmic trafficking of the Nipah virus matrix protein is important for viral budding. PLoS Pathog. 2010;6(11):e1001186. | |
Mayo MA. A summary of taxonomic changes recently approved by ICTV. Arch Virol. 2002;147(8):1655–1663. | |
Harcourt BH, Tamin A, Halpin K, et al. Molecular characterization of the polymerase gene and genomic termini of Nipah virus. Virology. 2001;287(1):192–201. | |
Harcourt BH, Lowe L, Tamin A, et al. Genetic characterization of Nipah virus, Bangladesh, 2004. Emerg Infect Dis. 2005;11(10):1594–1597. | |
Lo MK, Lowe L, Hummel KB, et al. Characterization of Nipah virus from outbreaks in Bangladesh, 2008–2010. Emerg Infect Dis. 2012; 18(2):248–255. | |
Clayton BA, Middleton D, Bergfeld J, et al. Transmission routes for Nipah virus from Malaysia and Bangladesh. Emerg Infect Dis. 2012; 18(12):1983–1993. | |
DeBuysscher BL, de Wit E, Munster VJ, Scott D, Feldmann H, Prescott J. Comparison of the pathogenicity of Nipah virus isolates from Bangladesh and Malaysia in the Syrian hamster. PLoS Negl Trop Dis. 2013;7(1):e2024. | |
Baseler L, de Wit E, Scott DP, Munster VJ, Feldmann H. Syrian hamsters (Mesocricetus auratus) oronasally inoculated with a Nipah virus isolate from Bangladesh or Malaysia develop similar respiratory tract lesions. Vet Pathol. 2015;52(1):38–45. | |
Lamb RA, Parks GD. Paramyxoviridae: the viruses and their replication. In: Fields BN, Knipe DM, Howley PM, editors. Fields’ Virology. 5th ed. Vol 1. Philadelphia, PA: Wolters Kluwer Heath/Lippincott Williams & Wilkins; 2007:1449–1496. | |
Yu F, Khairullah NS, Inoue S, et al. Serodiagnosis using recombinant nipah virus nucleocapsid protein expressed in Escherichia coli. J Clin Microbiol. 2006;44(9):3134–3138. | |
Foord AJ, White JR, Colling A, Heine HG. Microsphere suspension array assays for detection and differentiation of Hendra and Nipah viruses. Biomed Res Int. 2013;2013:289295. | |
Guillaume V, Lefeuvre A, Faure C, et al. Specific detection of Nipah virus using real-time RT-PCR (TaqMan). J Virol Methods. 2004; 120(2):229–237. | |
Patch JR, Han Z, McCarthy SE, et al. The YPLGVG sequence of the Nipah virus matrix protein is required for budding. Virol J. 2008;5:137. | |
Lamp B, Dietzel E, Kolesnikova L, et al. Nipah virus entry and egress from polarized epithelial cells. J Virol. 2013;87(6):3143–3154. | |
de Swart RL, Yüksel S, Osterhaus AD. Relative contributions of measles virus hemagglutinin- and fusion protein-specific serum antibodies to virus neutralization. J Virol. 2005;79(17):11547–11551. | |
Tamin A, Harcourt BH, Ksiazek TG, Rollin PE, Bellini WJ, Rota PA. Functional properties of the fusion and attachment glycoproteins of Nipah virus. Virology. 2002;296(1):190–200. | |
Prescott J, de Wit E, Feldmann H, Munster VJ. The immune response to Nipah virus infection. Arch Virol. 2012;157(9):1635–1641. | |
Diederich S, Sauerhering L, Weis M, et al. Activation of the Nipah virus fusion protein in MDCK cells is mediated by cathepsin B within the endosome-recycling compartment. J Virol. 2012;86(7):3736–3745. | |
Porotto M, Carta P, Deng Y, et al. Molecular determinants of antiviral potency of paramyxovirus entry inhibitors. J Virol. 2007;81(19):10567–10574. | |
Pernet O, Pohl C, Ainouze M, Kweder H, Buckland R. Nipah virus entry can occur by macropinocytosis. Virology. 2009;395(2):298–311. | |
Liu Q, Stone JA, Bradel-Tretheway B, et al. Unraveling a three-step spatiotemporal mechanism of triggering of receptor-induced Nipah virus fusion and cell entry. PLoS Pathog. 2013;9(11):e1003770. | |
Maisner A, Neufeld J, Weingartl H. Organ- and endotheliotropism of Nipah virus infections in vivo and in vitro. Thromb Haemost. 2009;102(6):1014–1023. | |
Wong KT, Shieh WJ, Kumar S, et al; Nipah Virus Pathology Working Group. Nipah virus infection: pathology and pathogenesis of an emerging paramyxoviral zoonosis. Am J Pathol. 2002;161(6):2153–2167. | |
Wong KT, Grosjean I, Brisson C, et al. A golden hamster model for human acute Nipah virus infection. Am J Pathol. 2003;163(5):2127–2137. | |
Li M, Embury-Hyatt C, Weingartl HM. Experimental inoculation study indicates swine as a potential host for Hendra virus. Vet Res. 2010; 41(3):33. | |
Liu Q, Bradel-Tretheway B, Monreal AI, et al. Nipah virus attachment glycoprotein stalk C-terminal region links receptor binding to fusion triggering. J Virol. 2015;89(3):1838–1850. | |
Maar D, Harmon B, Chu D, et al. Cysteines in the stalk of the nipah virus G glycoprotein are located in a distinct subdomain critical for fusion activation. J Virol. 2012;86(12):6632–6642. | |
Bowden TA, Crispin M, Harvey DJ, et al. Crystal structure and carbohydrate analysis of Nipah virus attachment glycoprotein: a template for antiviral and vaccine design. J Virol. 2008;82(23):11628–11636. | |
Bowden TA, Crispin M, Harvey DJ, Jones EY, Stuart DI. Dimeric architecture of the Hendra virus attachment glycoprotein: evidence for a conserved mode of assembly. J Virol. 2010;84(12):6208–6217. | |
Bossart KN, Crameri G, Dimitrov AS, et al. Receptor binding, fusion inhibition, and induction of cross-reactive neutralizing antibodies by a soluble G glycoprotein of Hendra virus. J Virol. 2005; 79(11):6690–6702. | |
Negrete OA, Levroney EL, Aguilar HC, et al. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature. 2005;436(7049):401–405. | |
Bonaparte MI, Dimitrov AS, Bossart KN, et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci U S A. 2005;102(30):10652–10657. | |
Tanimura N, Imada T, Kashiwazaki Y, et al. Reactivity of anti-Nipah virus monoclonal antibodies to formalin-fixed, paraffin-embedded lung tissues from experimental Nipah and Hendra virus infections. J Vet Med Sci. 2004;66(10):1263–1266. | |
Tanimura N, Imada T, Kashiwazaki Y, Shahirudin S, Sharifah SH, Aziz AJ. Monoclonal antibody-based immunohistochemical diagnosis of Malaysian Nipah virus infection in pigs. J Comp Pathol. 2004; 131(2–3):199–206. | |
Rockx B, Bossart KN, Feldmann F, et al. A novel model of lethal Hendra virus infection in African green monkeys and the effectiveness of ribavirin treatment. J Virol. 2010;84(19):9831–9839. | |
Pernet O, Wang YE, Lee B. Henipavirus receptor usage and tropism. Curr Top Microbiol Immunol. 2012;359:59–78. | |
Bochenek ML, Dickinson S, Astin JW, Adams RH, Nobes CD. Ephrin-B2 regulates endothelial cell morphology and motility independently of Eph-receptor binding. J Cell Sci. 2010;123(Pt 8):1235–1246. | |
Yu G, Luo H, Wu Y, Wu J. Ephrin B2 induces T cell costimulation. J Immunol. 2003;171(1):106–114. | |
Halpin K, Bankamp B, Harcourt BH, Bellini WJ, Rota PA. Nipah virus conforms to the rule of six in a minigenome replication assay. J Gen Virol. 2004;85(Pt 3):701–707. | |
Chan YP, Koh CL, Lam SK, Wang LF. Mapping of domains responsible for nucleocapsid protein-phosphoprotein interaction of Henipaviruses. J Gen Virol. 2004;85(Pt 6):1675–1684. | |
Wang LF, Michalski WP, Yu M, et al. A novel P/V/C gene in a new member of the Paramyxoviridae family, which causes lethal infection in humans, horses, and other animals. J Virol. 1998; 72(2):1482–1490. | |
Harcourt BH, Tamin A, Ksiazek TG, et al. Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology. 2000;271(2):334–349. | |
Kulkarni S, Volchkova V, Basler CF, Palese P, Volchkov VE, Shaw ML. Nipah virus edits its P gene at high frequency to express the V and W proteins. J Virol. 2009;83(8):3982–3987. | |
Lo MK, Harcourt BH, Mungall BA, et al. Determination of the henipavirus phosphoprotein gene mRNA editing frequencies and detection of the C, V and W proteins of Nipah virus in virus-infected cells. J Gen Virol. 2009;90(Pt 2):398–404. | |
Rota PA, Lo MK. Molecular virology of the henipaviruses. Curr Top Microbiol Immunol. 2012;359:41–58. | |
Shaw ML, García-Sastre A, Palese P, Basler CF. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J Virol. 2004;78(11):5633–5641. | |
Rodriguez JJ, Parisien JP, Horvath CM. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J Virol. 2002;76(22):11476–11483. | |
Rodriguez JJ, Cruz CD, Horvath CM. Identification of the nuclear export signal and STAT-binding domains of the Nipah virus V protein reveals mechanisms underlying interferon evasion. J Virol. 2004; 78(10):5358–5367. | |
Shaw ML, Cardenas WB, Zamarin D, Palese P, Basler CF. Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and toll-like receptor 3-triggered signaling pathways. J Virol. 2005;79(10):6078–6088. | |
Childs K, Stock N, Ross C, et al. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology. 2007;359(1):190–200. | |
Parisien JP, Bamming D, Komuro A, et al. A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J Virol. 2009;83(14):7252–7260. | |
Yamaguchi M, Kitagawa Y, Zhou M, Itoh M, Gotoh B. An anti-interferon activity shared by paramyxovirus C proteins: inhibition of Toll-like receptor 7/9-dependent alpha interferon induction. FEBS Lett. 2014;588(1):28–34. | |
Lo MK, Peeples ME, Bellini WJ, Nichol ST, Rota PA, Spiropoulou CF. Distinct and overlapping roles of Nipah virus P gene products in modulating the human endothelial cell antiviral response. PLoS One. 2012;7(10):e47790. | |
Mathieu C, Guillaume V, Volchkova VA, et al. Nonstructural Nipah virus C protein regulates both the early host proinflammatory response and viral virulence. J Virol. 2012;86(19):10766–10775. | |
Sleeman K, Bankamp B, Hummel KB, Lo MK, Bellini WJ, Rota PA. The C, V and W proteins of Nipah virus inhibit minigenome replication. J Gen Virol. 2008;89(Pt 5):1300–1308. | |
Park MS, Shaw ML, Muñoz-Jordan J, et al. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J Virol. 2003;77(2):1501–1511. | |
Yoneda M, Fujita K, Sato H, Kai C. Reverse genetics of Nipah virus to probe viral pathogenicity. Methods Mol Biol. 2009;515:329–337. | |
Yoneda M, Guillaume V, Sato H, et al. The nonstructural proteins of Nipah virus play a key role in pathogenicity in experimentally infected animals. PLoS One. 2010;5(9):e12709. | |
Virtue ER, Marsh GA, Wang LF. Interferon signaling remains functional during henipavirus infection of human cell lines. J Virol. 2011;85(8):4031–4034. | |
Chong HT, Kamarulzaman A, Tan CT, et al. Treatment of acute Nipah encephalitis with ribavirin. Ann Neurol. 2001;49(6):810–813. | |
Chong HT, Kunjapan SR, Thayaparan T, et al. Nipah encephalitis outbreak in Malaysia, clinical features in patients from Seremban. Can J Neurol Sci. 2002;29(1):83–87. | |
Wong KT, Tan CT. Clinical and pathological manifestations of human henipavirus infection. Curr Top Microbiol Immunol. 2012; 359:95–104. | |
O’Sullivan JD, Allworth AM, Paterson DL, et al. Fatal encephalitis due to novel paramyxovirus transmitted from horses. Lancet. 1997; 349(9045):93–95. | |
Tan CT, Goh KJ, Wong KT, et al. Relapsed and late-onset Nipah encephalitis. Ann Neurol. 2002;51(6):703–708. | |
Mathieu C, Pohl C, Szecsi J, et al. Nipah virus uses leukocytes for efficient dissemination within a host. J Virol. 2011;85(15):7863–7871. | |
Stachowiak B, Weingartl HM. Nipah virus infects specific subsets of porcine peripheral blood mononuclear cells. PLoS One. 2012; 7(1):e30855. | |
Wong SC, Ooi MH, Wong MN, Tio PH, Solomon T, Cardosa MJ. Late presentation of Nipah virus encephalitis and kinetics of the humoral immune response. J Neurol Neurosurg Psychiatry. 2001; 71(4):552–554. | |
Chan KP, Rollin PE, Ksiazek TG, et al. A survey of Nipah virus infection among various risk groups in Singapore. Epidemiol Infect. 2002;128(1):93–98. | |
Middleton DJ, Weingartl HM. Henipaviruses in their natural animal hosts. Curr Top Microbiol Immunol. 2012;359:105–121. | |
Hooper PT, Ketterer PJ, Hyatt AD, Russell GM. Lesions of experimental equine morbillivirus pneumonia in horses. Vet Pathol. 1997; 34(4):312–322. | |
Rogers RJ, Douglas IC, Baldock FC, et al. Investigation of a second focus of equine morbillivirus infection in coastal Queensland. Aust Vet J. 1996;74(3):243–244. | |
Black PF, Cronin JP, Morrissy CJ, Westbury HA. Serological examination for evidence of infection with Hendra and Nipah viruses in Queensland piggeries. Aust Vet J. 2001;79(6):424–426. | |
Middleton DJ, Westbury HA, Morrissy CJ, et al. Experimental Nipah virus infection in pigs and cats. J Comp Pathol. 2002;126(2–3):124–136. | |
Weingartl HM, Berhane Y, Caswell JL, et al. Recombinant nipah virus vaccines protect pigs against challenge. J Virol. 2006; 80(16):7929–7938. | |
Weingartl HM, Berhane Y, Czub M. Animal models of henipavirus infection: a review. Vet J. 2009;181(3):211–220. | |
Middleton D, Pallister J, Klein R, et al. Hendra virus vaccine, a one health approach to protecting horse, human, and environmental health. Emerg Infect Dis. 2014;20(3):372–379. | |
Westbury HA, Hooper PT, Brouwer SL, Selleck PW. Susceptibility of cats to equine morbillivirus. Aust Vet J. 1996;74(2):132–134. | |
Westbury HA, Hooper PT, Selleck PW, Murray PK. Equine morbillivirus pneumonia: susceptibility of laboratory animals to the virus. Aust Vet J. 1995;72(7):278–279. | |
Mungall BA, Middleton D, Crameri G, et al. Feline model of acute nipah virus infection and protection with a soluble glycoprotein-based subunit vaccine. J Virol. 2006;80(24):12293–12302. | |
Mungall BA, Middleton D, Crameri G, et al. Vertical transmission and fetal replication of Nipah virus in an experimentally infected cat. J Infect Dis. 2007;196(6):812–816. | |
Mills JN, Alim AN, Bunning ML, et al. Nipah virus infection in dogs, Malaysia, 1999. Emerg Infect Dis. 2009;15(6):950–952. | |
Bossart KN, Zhu Z, Middleton D, et al. A neutralizing human monoclonal antibody protects against lethal disease in a new ferret model of acute nipah virus infection. PLoS Pathog. 2009; 5(10):e1000642. | |
Pallister J, Middleton D, Crameri G, et al. Chloroquine administration does not prevent Nipah virus infection and disease in ferrets. J Virol. 2009;83(22):11979–11982. | |
Pallister J, Middleton D, Wang LF, et al. A recombinant Hendra virus G glycoprotein-based subunit vaccine protects ferrets from lethal Hendra virus challenge. Vaccine. 2011;29(34):5623–5630. | |
Pallister JA, Klein R, Arkinstall R, et al. Vaccination of ferrets with a recombinant G glycoprotein subunit vaccine provides protection against Nipah virus disease for over 12 months. Virol J. 2013; 10:237. | |
Mire CE, Versteeg KM, Cross RW, et al. Single injection recombinant vesicular stomatitis virus vaccines protect ferrets against lethal Nipah virus disease. Virol J. 2013;10:353. | |
Marsh GA, Virtue ER, Smith I, et al. Recombinant Hendra viruses expressing a reporter gene retain pathogenicity in ferrets. Virol J. 2013;10:95. | |
Williamson MM, Hooper PT, Selleck PW, Westbury HA, Slocombe RF. A guinea-pig model of Hendra virus encephalitis. J Comp Pathol. 2001;124(4):273–279. | |
Dhondt KP, Horvat B. Henipavirus infections: lessons from animal models. Pathogens. 2013;2(2):264–287. | |
Dups J, Middleton D, Yamada M, et al. A new model for Hendra virus encephalitis in the mouse. PLoS One. 2012;7(7):e40308. | |
Valbuena G, Halliday H, Borisevich V, Goez Y, Rockx B. A human lung xenograft mouse model of Nipah virus infection. PLoS Pathog. 2014;10(4):e1004063. | |
Yun T, Park A, Hill TE, et al. Efficient reverse genetics reveals genetic determinants of budding and fusogenic differences between Nipah and Hendra viruses and enables real-time monitoring of viral spread in small animal models of henipavirus infection. J Virol. 2015;89(2):1242–1253. | |
Defang GN, Khetawat D, Broder CC, Quinnan GV. Induction of neutralizing antibodies to Hendra and Nipah glycoproteins using a Venezuelan equine encephalitis virus in vivo expression system. Vaccine. 2010;29(2):212–220. | |
Kong D, Wen Z, Su H, et al. Newcastle disease virus-vectored Nipah encephalitis vaccines induce B and T cell responses in mice and long-lasting neutralizing antibodies in pigs. Virology. 2012; 432(2):327–335. | |
Kurup D, Wirblich C, Feldmann H, Marzi A, Schnell MJ. Rhabdovirus- based vaccine platforms against henipaviruses. J Virol. 2015;89(1):144–154. | |
Guillaume V, Contamin H, Loth P, et al. Nipah virus: vaccination and passive protection studies in a hamster model. J Virol. 2004; 78(2):834–840. | |
Rockx B, Brining D, Kramer J, et al. Clinical outcome of henipavirus infection in hamsters is determined by the route and dose of infection. J Virol. 2011;85(15):7658–7671. | |
de Wit E, Munster VJ. Animal models of disease shed light on Nipah virus pathogenesis and transmission. J Pathol. 2015;235(2):196–205. | |
de Wit E, Prescott J, Falzarano D, et al. Foodborne transmission of nipah virus in Syrian hamsters. PLoS Pathog. 2014;10(3):e1004001. | |
de Wit E, Bushmaker T, Scott D, Feldmann H, Munster VJ. Nipah virus transmission in a hamster model. PLoS Negl Trop Dis. 2011; 5(12):e1432. | |
Guillaume V, Wong KT, Looi RY, et al. Acute Hendra virus infection: Analysis of the pathogenesis and passive antibody protection in the hamster model. Virology. 2009;387(2):459–465. | |
Munster VJ, Prescott JB, Bushmaker T, et al. Rapid Nipah virus entry into the central nervous system of hamsters via the olfactory route. Sci Rep. 2012;2:736. | |
Marianneau P, Guillaume V, Wong T, et al. Experimental infection of squirrel monkeys with nipah virus. Emerg Infect Dis. 2010; 16(3):507–510. | |
Geisbert TW, Daddario-DiCaprio KM, Hickey AC, et al. Development of an acute and highly pathogenic nonhuman primate model of Nipah virus infection. PLoS One. 2010;5(5):e10690. | |
Mire CE, Geisbert JB, Agans KN, et al. A recombinant Hendra virus G glycoprotein subunit vaccine protects nonhuman primates against Hendra virus challenge. J Virol. 2014;88(9):4624–4631. | |
Johnston SC, Briese T, Bell TM, et al. Detailed analysis of the African green monkey model of Nipah virus disease. PLoS One. 2015;10(2):e0117817. | |
Yoneda M, Georges-Courbot MC, Ikeda F, et al. Recombinant measles virus vaccine expressing the Nipah virus glycoprotein protects against lethal Nipah virus challenge. PLoS One. 2013;8(3):e58414. | |
Bossart KN, Rockx B, Feldmann F, et al. A Hendra virus G glycoprotein subunit vaccine protects African green monkeys from Nipah virus challenge. Sci Transl Med. 2012;4(146):146ra107. | |
McEachern JA, Bingham J, Crameri G, et al. A recombinant subunit vaccine formulation protects against lethal Nipah virus challenge in cats. Vaccine. 2008;26(31):3842–3852. | |
Ploquin A, Szécsi J, Mathieu C, et al. Protection against henipavirus infection by use of recombinant adeno-associated virus-vector vaccines. J Infect Dis. 2013;207(3):469–478. | |
DeBuysscher BL, Scott D, Marzi A, Prescott J, Feldmann H. Single-dose live-attenuated Nipah virus vaccines confer complete protection by eliciting antibodies directed against surface glycoproteins. Vaccine. 2014;32(22):2637–2644. | |
Lo MK, Bird BH, Chattopadhyay A, et al. Single-dose replication-defective VSV-based Nipah virus vaccines provide protection from lethal challenge in Syrian hamsters. Antiviral Res. 2014;101:26–29. | |
Prescott J, DeBuysscher BL, Feldmann F, et al. Single-dose live-attenuated vesicular stomatitis virus-based vaccine protects African green monkeys from Nipah virus disease. Vaccine. 2015; 33(24):2823–2829. | |
Freiberg AN, Worthy MN, Lee B, Holbrook MR. Combined chloroquine and ribavirin treatment does not prevent death in a hamster model of Nipah and Hendra virus infection. J Gen Virol. 2010;91(Pt 3):765–772. | |
Georges-Courbot MC, Contamin H, Faure C, et al. Poly(I)-poly(C12U) but not ribavirin prevents death in a hamster model of Nipah virus infection. Antimicrob Agents Chemother. 2006;50(5):1768–1772. | |
Zhu Z, Bossart KN, Bishop KA, et al. Exceptionally potent cross-reactive neutralization of Nipah and Hendra viruses by a human monoclonal antibody. J Infect Dis. 2008;197(6):846–853. | |
Geisbert TW, Mire CE, Geisbert JB, et al. Therapeutic treatment of Nipah virus infection in nonhuman primates with a neutralizing human monoclonal antibody. Sci Transl Med. 2014;6(242):242ra82. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.