Back to Journals » Cancer Management and Research » Volume 13
HCP5 Promotes Proliferation and Contributes to Cisplatin Resistance in Gastric Cancer Through miR-519d/HMGA1 Axis
Received 2 November 2020
Accepted for publication 31 December 2020
Published 27 January 2021 Volume 2021:13 Pages 787—794
DOI https://doi.org/10.2147/CMAR.S289997
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Professor Seema Singh
Zhu Zhang, Huahong Wang
Department of Gastroenterology, Peking University First Hospital, Beijing 100034, People’s Republic of China
Correspondence: Huahong Wang
Department of Gastroenterology, Peking University First Hospital, Beijing 100034, People’s Republic of China
Tel +86-10-83572301
Email [email protected]
Introduction: The long-non-coding RNA HCP5 (HLA complex P5) has been extensively linked to the ability of cancer cells to resist chemotherapeutic interventions. Here, we investigated the role of HCP5 in gastric cancer (GC) which to-date has been poorly characterized. Our results indicated that HCP5 expression was up-regulated in GC cells.
Methods: HCP5, miR-519d, and high mobility group A1 (HMGA1) expression levels in GC cells were measured using quantitative real-time PCR (qRT-PCR) and Western blot analysis. Drug sensitivity and apoptosis of tumor cells were assessed using cell counting kit-8, flow cytometry, and caspase activity assay. Bioinformatics and luciferase reporter assays were employed for analyzing the interactions between HCP5, miR-519d, and HMGA1.
Results: HCP5 knockdown suppressed proliferation and weakened the resistance to cisplatin (DDP) of GC cells. miR-519d was down-regulated in GC cells and sponged by HCP5. HMGA1 was directly inhibited by miR-519d and its expression was up-regulated in GC cells. HCP5 exacerbated the resistance to cisplatin of GC cells in vitro by enhancing HMGA1 expression via sponging miR-519d.
Conclusion: In summary, HCP5 promoted proliferation and contributed to DDP resistance in GC cells through miR-519d/HMGA1 axis.
Keywords: lncRNA HCP5, gastric cancer, cisplatin, miR-519d, HMGA1
Introduction
Gastric cancer (GC) is one of the most prevalent malignancies and the secondary leading cause of cancer-related deaths worldwide.1 According to the latest global cancer statistics, the deaths of GC are up to 782,685 in 2018, accounting for 8.2% among all cancer deaths in that year.2 Surgical resection is the first choice of treatment for patients with early-stage GC. However, a large proportion of the patients are diagnosed at an advanced stage, and a combination of surgical resection, radiotherapy, and diverse chemotherapy are frequently used for patients with advanced-stage GC.3,4 Due to its high recurrence rate and high metastasis rate, the overall survival rate of patients with GC still remains poor.5 Hence, safe and effective drugs capable of preventing GC are urgently required to improve the therapeutic outcomes.
RNAs that lack coding function (non-coding) regulate gene expression in a range of cell types. Specifically, these RNAs that exceed 200 nts in length are termed lncRNAs, the dysregulation of which have been implicated in tumor growth and formation.6,7 In various cancers, lncRNAs can promote cancer formation or can act as inhibitors of metastatic processes. Many lncRNAs contribute to tumor prognosis, cancer diagnosis,8 and drug resistance,9 such as sunitinib resistance in renal cancer10 and DDP resistance in GC.11 LncRNA HLA complex P5 (HCP5) is primarily found expressed in immune system cells. Recently, lncRNA HCP5 has been reported to promote the growth, metastasis, and drug resistance in various cancers, such as breast cancer,12 pancreatic cancer,13 and lung cancer.14 Nevertheless, the function and mechanism of HCP5 in GC tumorigenesis are still elusive.
MicroRNAs (miRNAs), also belonging to non-coding RNAs (ncRNAs), emerge as a type of endogenous ncRNAs with 19–24 nucleotides, and can bind to the 3ʹ untranslated region (3ʹ-UTR) of target gene mRNA to further modulate the target gene expression.15,16 In addition, miRNAs were reported to actively take part in the regulation of various cellular processes, including tumorigenesis and chemoresistance.17 Previous studies report that miR-519d acted as a tumor suppressor in GC18 and overcame DDP resistance in ovarian cancer cells.19 Furthermore, high mobility group A1 (HMGA1) has been reported to promote proliferation and contribute to DDP resistance in various tumors, such as ovarian cancer and bladder cancer.20,21 The bioinformatics analysis predicted HCP5 and HMGA1 could be targeted via miR-519d. Therefore, we hypothesized that HCP5 might participate in GC progression via modulating miR-519d and HMGA1.
In this work, we investigated the functional role of HCP5 in tumor progression and DDP resistance in GC cells. Additionally, we explored the potential competing endogenous RNA (ceRNA) mechanism of HCP5/miR-519d/HMGA1.
Materials and Methods
Cell Culture and Transfection
Normal gastric cell line (GES-1) and GC cell lines (NCI-N87, MKN-45, SNU-1, and KATO-3) from Shanghai Cell Bank (Shanghai, China) were cultured in Dulbecco’s Modified Eagle Medium/Ham’s Nutrient mixture F12 (DMEM/F12, Gibco, Grand Island, NY, USA) containing 10% FBS (HyClone, Logan, UT, USA) in 5% CO2 at 37°C.
siRNA targeting HCP5 (si-HCP5#1, si-HCP5#2, si-HCP5 #3), si-control (si-con), miR-519d, miR-con, and anti-miR-519d were acquired from GenePharma (Shanghai, China). The sequence of HCP5 was amplified and inserted into the pcDNA-3.1 vector (Invitrogen, Carlsbad, CA, USA) to create pcDNA-HCP5 (oe-HCP5). Cell transfection was conducted by use of Lipofectamine 2000 (Invitrogen), in the light of the manufacturer’s recommendations.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA extraction was conducted following the kit instructions, and Nanodrop-spectrophotometer was employed to measure the RNA purity and concentration. Then, based on the manufacturer’s protocol, complementary DNA (cDNA) was synthesized from 1 µg of total RNA with the PrimeScript-RT Kit (Madison, WI, USA), and then the SYBR® Premix-Ex-Taq™ (Takara, TX, USA) and ABI7300 system were used to perform PCR. All fluorescence data were converted into relative quantification, with U6 and GAPDH treated as the internal reference for miRNA, and mRNA, respectively. The 2−ΔΔCt method was used to analyze the data.
Cell Proliferation and Drug Sensitivity Assay
Cell counting kit-8 (CCK-8) assay was performed to test the rate of cell proliferation. In brief, transfected SNU-1 or KATO-3 cells were plated in the 96-well plates at a density of 1 × 103 cells/well and cultured in an incubator at indicated time. Following that, cell viability was determined by using Cell Counting Kit-8 (Keygen, China).
The GC cells were planted into 96-well plates, followed by incubation with different concentrations (0.5, 1, 2, 4, 8, 16, 32 μM) of DDP. At 48 h after cultivation, the cells were incubated with CCK-8 solution (Beyotime, Shanghai, China) for 2 h and then tested for the absorbance at 450 nm using a microplate reader.
EdU Proliferation Assay
Five-ethynyl-2ʹ-deoxyuridine (EdU) incorporation assays were carried out as described previously.22 Briefly, 5 x 103 cells per well were grown in a 96-well dish and processed with the EdU labeling kit (RiboBio, Guangzhou, China).
Flow Cytometric Analysis
Annexin V-FITC/PI Apoptosis Detection Kit was bought from KeyGEN Biotech (Nanjing, China). Briefly, cells harvested during logarithmic growth were suspended in Annexin-binding buffer. Then, the cells were stained using Annexin V/FITC and PI solution and incubated in the dark at room temperature for 15 min. Finally, cells were detected by flow cytometry analysis.
Caspase Activity Assay
The activities of caspase-3 and caspase-9 in transfected SNU-1 and KATO-3 cells treated with 4 μM DDP were detected using a colorimetric assay kit (R&D Systems Inc., Minneapolis, MN, USA) according to the manufacturer’s instruction.
Luciferase Reporter Assay
The dual-luciferase activity detection kit was purchased from Promega Corporation (Promega, Madison, WI, USA), and luciferase reporter vectors were synthesized by Promega Corporation. Reporter gene plasmids for wild-type (WT) and mutant-type (MUT) HCP5 and HMGA1-3ʹUTR were constructed and co-transfected into GC cells with miR-519d and miR-con, respectively. After 48 h of transfection, for each group, firefly luciferase and renilla luciferase activity were detected with a microplate reader.
Western Blotting
The protein expression levels of HMGA1 were evaluated in SNU-1 and KATO-3 cells as previously described.23 The following antibodies were used: anti-rabbit HMGA1 (Abcam, Cambridge, UK) (1:1000), anti-rabbit GAPDH ((Cell Signaling Technology, Inc., Danvers, MA, USA)) (1:1000), anti-rabbit HRP-linked secondary IgG antibody (Cell Signaling) (1:2000).
Statistical Analysis
Mean ± standard deviation was the expression form of all data. One-way ANOVA was performed for comparing the data difference among 3 or more groups, and t-test was utilized for comparing the difference between two groups. P < 0.05 represented that differences were of statistical significance.
Results
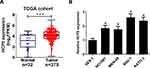
HCP5 Was Overexpressed in GC Cells
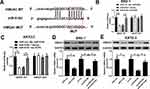
To investigate the biological role of lncRNA HCP5 in GC, we analyzed the data of TCGA Stomach Adenocarcinoma (STAD) and found that HCP5 was significantly up-regulated in GC tumor tissues (Figure 1A). To further confirm the level of HCP5 in GC cells, we analyzed HCP5 expression in GC cell lines (NCI-N87, MKN-45, SNU-1, and KATO-3) and normal gastric cell line (GES-1). HCP5 was significantly increased in GC cells compared with GES-1 cells, especially in SNU-1 and KATO-3 cells (Figure 1B). Collectively, HCP5 up-regulation may be implicated with GC tumorigenesis.
HCP5 Silencing Inhibited Cell Proliferation and Enhanced DDP Sensitivity in GC Cells
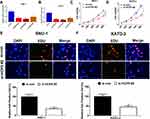
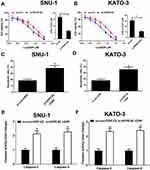
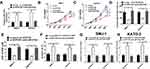
To confirm the functional role of HCP5 in SNU-1 and KATO-3 cells, the cells were transfected with si-con, si-HCP5#1, si-HCP5#2, or si-HCP5#3, and the transfection efficiency was identified using qRT-PCR. The results revealed that the expression of HCP5 was markedly reduced in si-HCP5 group (Figure 2A and B). To investigate the effect of HCP5 on cell proliferation in GC cells, CCK-8 and EdU proliferation assays were performed in SNU-1 and KATO-3 cells transfected with si-con or si-HCP5 #2. As expected, HCP5 knockdown suppressed cell proliferation in SNU-1 and KATO-3 cells (Figure 2C–F). To further explore the effect of HCP5 on DDP resistance in GC cells, si-con or si-HCP5 #2 transfected SNU-1 and KATO-3 cells were treated with increasing doses (0.5, 1, 2, 4, 8, 16, 32 μM) of DDP and then subjected to CCK-8 assay. The results indicated that HCP5 silencing could sensitize SNU-1 and KATO-3 cells to DDP, evidenced by the reduced IC50 value (Figure 3A and B). In parallel, knockdown of HCP5 promoted the apoptosis of SNU-1 and KATO-3 cells in the presence of DDP, as determined by flow cytometry analysis and Caspase activity assay (Figure 3C–F). Collectively, knockdown of HCP5 suppressed proliferation and improved DDP sensitivity in GC cells.
HCP5 Acts as a Sponge of miR-519d in GC Cells
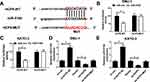
To investigate the downstream targets of HCP5, we performed the online bioinformatics database starBase 3.0 prediction, which indicated that HCP5 had the binding site for miR-519d (Figure 4A). Dual-luciferase reporter gene assay indicated that miR-519d mimics could inhibit the luciferase activity of HCP5-WT, while it did not significantly affect the luciferase activity of HCP5-MUT (Figure 4B and C). Moreover, qRT-PCR showed that HCP5 overexpression could inhibit miR-519d expression, while si-HCP5 #2 promoted its expression (Figure 4D and E). To sum up, HCP5 directly targeted miR-519d and negatively regulated its expression in GC cells.
HCP5 Knockdown Suppresses Proliferation and Facilitates DDP Sensitivity of GC Cells Through Adsorbing miR-519d
The foregoing experiments showed that HCP5 could adsorb miR-519d, thereby inhibiting its expression. To investigate the effect of HCP5 on cell proliferation by adsorbing miR-519d, SNU-1, and KATO-3 cells were transfected with si-HCP5 #2 or si-HCP5 #2+anti-miR-519d. qRT-PCR verified the success of the transfection (Figure 5A). CCK-8 and EdU proliferation assays revealed that miR-519d inhibition weakened the suppressive effect of si-HCP5 #2 on cell proliferation in SNU-1 and KATO-3 cells (Figure 5B–D). Subsequently, drug sensitivity, flow cytometry, and caspase activity assays manifested that miR-519d inhibition debilitated the enhancive effect of si-HCP5 #2 on DDP sensitivity and apoptosis (Figure 5E–H). These indicated that HCP5 silencing could inhibit GC proliferation and enhance DDP sensitivity by regulating miR-519d expression.
HCP5 Promotes HMGA1 Expression in GC Cells Through Sponging miR-519d
Public database starBase 3.0 prediction shows the binding site between miR-519d and HMGA1 (Figure 6A). Consistently, dual-luciferase reporter gene assay suggested that the luciferase activity of HMGA1-WT construct was significantly decreased after miR-519d transfection, which was blocked by up-regulation of HCP5. However, there was no significant change in the luciferase activity of HMGA1-MUT in all groups (Figure 6B and C). miR-519d mimics or HCP5 silencing could suppress HMGA1 expression, while miR-519d inhibitors reversed the inhibitory effect of si-HCP5 #2 on HMGA1 expression (Figure 6D and E). All the above-mentioned findings supported that HCP5 elevated HMGA1 expression in GC cells through adsorbing miR-519d.
Discussion
During the past several decades, in spite of the remarkable progresses in diagnostic and therapeutic methods of GC, the long-term survival of GC patients is unsatisfactory because of cancer recurrence and chemoresistance. Thus, deeply investigating key signaling pathway driving GC progression and chemoresistance is crucial for discovering new therapeutic targets and strategies of GC.
Recent evidences have shown that lncRNA participates in multistep of cancer development by affecting key genes modulating cell proliferation, apoptosis, and invasion. HCP5, a novel identified lncRNA, is overexpressed in many cancers.12–14 Moreover, emerging evidence suggested that HCP5 was implicated in chemoresistance.13,24 Particularly, HCP5 contributed to DDP resistance in breast cancer cells through inhibiting phosphatase and tensin homolog (PTEN) expression.25 Nevertheless, until now, the HCP5 function in GC is still unclear. In this research, real-time PCR analysis results showed that GC cells exhibited higher HCP5 levels than controls. Additionally, HCP5 knockdown suppressed cell proliferation and DPP resistance in GC cells, suggesting the tight relationship between HCP5 and GC development and chemoresistance.
Mounting researches proposed that the lncRNAs could adsorb miRNA to set free miRNA’s target genes, thus participating in multiple pathogenic processes especially in cancer. Herein, Dual-luciferase assay results verified that miR-519d could bind to HCP5 directly. HCP5 knockdown increased the miR-519d level, while miR-519d overexpression had no influences on HCP5 expression. As expected, GC cells displayed lower miR-519d levels than controls. Functional results revealed HCP5 knockdown suppressed cell proliferation and overcame DDP resistance in GC cells, while miR-519d inhibitor could offset these effects caused by HCP5 ablation, suggesting HCP5 modulated GC development and chemoresistance in a miR-519d dependent manner. Moreover, HMGA1 was further verified as a miR-519d target in this work. HMGA1 is an oncogene, which is required for various types of tumor development by modulating cancer cell apoptosis, growth, and invasion.26–29 In addition, HMGA1 has been reported to confer DDP resistance in many tumors, including ovarian cancer and bladder cancer.20,21 In this work, consistently, we proved that miR-519d could suppress the expression of HMGA1 via targeting it, and HCP5 could positively regulate HMGA1, which could be reversed by miR-519d. Therefore, HCP5 could elevate HMGA1 expression through sponging miR-519d in GC. These demonstrations partly explained the mechanism by which HCP5 participated in GC progression and DDP chemoresistance.
In summary, our findings disclose an unexplored HCP5/miR-519d/HMGA1 axis in regulating gastric cancer progression and DDP resistance. Therefore, it may provide a promising therapeutic option for gastric cancer treatment.
Disclosure
No competing financial interests exist. The authors report no conflicts of interest for this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Stessin AM, Sison C, Schwartz A, et al. Does adjuvant radiotherapy benefit patients with diffuse-type gastric cancer? Results from the surveillance, epidemiology, and end results database. Cancer. 2015;120(22):3562–3568. doi:10.1002/cncr.28913
4. Nakayama Y, Mimura K, Kua LF, et al. Immune suppression caused by PD-L2 expression on tumor cells in gastric cancer. Gastric Cancer. 2020;23:961–973. doi:10.1007/s10120-020-01079-z
5. Sgambato D, Miranda A, Romano L, Romano M. Gut microbiota and gastric disease. Minerva Gastroenterol Dietol. 2017;63(4):345–354. doi:10.23736/S1121-421X.17.02380-7
6. Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26(2):155–165. doi:10.1038/modpathol.2012.160
7. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391. doi:10.1158/2159-8290.CD-11-0209
8. Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol. 2013;34(2):613–620. doi:10.1007/s13277-013-0658-6
9. Wang H, Guan Z, He K, Qian J, Cao J, Teng L. LncRNA UCA1 in anti-cancer drug resistance. Oncotarget. 2017;8(38):64638–64650. doi:10.18632/oncotarget.18344
10. Qu L, Ding J, Cheng C, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29(5):653–668. doi:10.1016/j.ccell.2016.03.004
11. Zhang Y, Song X, Wang X, Hu J, Jiang L. Silencing of LncRNA HULC enhances chemotherapy induced apoptosis in human gastric cancer. J Med Biochem. 2016;35(2):137–143. doi:10.1515/jomb-2015-0016
12. Wang L, Luan T, Zhou S, Lin J, Yang Y. LncRNA HCP5 promotes triple negative breast cancer progression as a ceRNA to regulate BIRC3 by sponging miR-219a-5p. Cancer Med. 2019;8(4):4389–4403. doi:10.1002/cam4.2335
13. Chen R, Xin G, Zhang X. Long non-coding RNA HCP5 serves as a ceRNA sponging miR-17-5p and miR-27a/b to regulate the pathogenesis of childhood obesity via the MAPK signaling pathway. J Pediatr Endocrinol Metab. 2019;32(12).
14. Jiang L, Wang R, Fang L, Ge X, Chen L. HCP5 is a SMAD3-responsive long non-coding RNA that promotes lung adenocarcinoma metastasis via miR-203/SNAI axis. Theranostics. 2019;9(9):2460–2474. doi:10.7150/thno.31097
15. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi:10.1038/nrg.2016.20
16. Ding X, Zheng Y, Wang Z, et al. Construction and investigation of lncRNA-associated ceRNA regulatory network in papillary thyroid cancer. Oncol Rep. 2018;39:3. doi:10.3892/or.2018.6335
17. Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi:10.1146/annurev-pathol-012513-104715
18. Yue H, Tang B, Zhao Y, et al. MIR-519d suppresses the gastric cancer epithelial-mesenchymal transition via Twist1 and inhibits Wnt/β-catenin signaling pathway. Am J Transl Res. 2017;9(8):3654–3664.
19. Pang Y, Mao H, Shen L, Zhao Z, Liu R, Liu P. MiR-519d represses ovarian cancer cell proliferation and enhances cisplatin-mediated cytotoxicity in vitro by targeting XIAP. Onco Targets Ther. 2014;587. doi:10.2147/OTT.S60289
20. Chen Y, Ren C, Yang L, et al. MicroRNA let‑7d‑5p rescues ovarian cancer cell apoptosis and restores chemosensitivity by regulating the p53 signaling pathway via HMGA1. Int J Oncol. 2019;54(5):1771–1784. doi:10.3892/ijo.2019.4731
21. Chen X, Liu M, Meng F, Sun B, Jin X, Jia C. The long noncoding RNA HIF1A-AS2 facilitates cisplatin resistance in bladder cancer. J Cell Biochem. 2019;120(1):243–252. doi:10.1002/jcb.27327
22. Zhang W, Kong G, Zhang J, Wang T, Ye L, Zhang X. MicroRNA-520b inhibits growth of hepatoma cells by targeting MEKK2 and cyclin D1. PLoS One. 2012;7(2):e31450. doi:10.1371/journal.pone.0031450
23. Wang J, Xu W, He Y, et al. LncRNA MEG3 impacts proliferation, invasion, and migration of ovarian cancer cells through regulating PTEN[J]. Inflamm Res. 2018;67:927–936. doi:10.1007/s00011-018-1186-z
24. Wu H, Liu B, Chen Z, Li G, Zhang Z. MSC-induced lncRNA HCP5 drove fatty acid oxidation through miR-3619-5p/AMPK/PGC1α/CEBPB axis to promote stemness and chemo-resistance of gastric cancer. Cell Death Dis. 2020;11(4):233. doi:10.1038/s41419-020-2426-z
25. Wu J, Chen H, Ye M, et al. Downregulation of long noncoding RNA HCP5 contributes to cisplatin resistance in human triple-negative breast cancer via regulation of PTEN expression. Biomed Pharmacother. 2019;115:108869. doi:10.1016/j.biopha.2019.108869
26. Ma H, Wang J, Shi J, Zhang W, Zhou D. LncRNA LINC00467 contributes to osteosarcoma growth and metastasis through regulating HMGA1 by directly targeting miR-217. Eur Rev Med Pharmacol Sci. 2020;24(11):5933–5945. doi:10.26355/eurrev_202006_21486
27. Wu Z, Wang W, Wang Y, et al. LINC00963Long noncoding RNA promotes breast cancer progression by functioning as a molecular sponge for microRNA-625 and thereby upregulating HMGA1. Cell Cycle. 2020;19(5):610–624. doi:10.1080/15384101.2020.1728024
28. Liu L, Zhang S, Hu L, Liu L, Guo W, Zhang J. HMGA1 participates in MHCC97H cell proliferation and invasion through the ILK/Akt/GSK3β signaling pathway. Mol Med Rep. 2017;16(6):9287–9294. doi:10.3892/mmr.2017.7820
29. Sekimoto N, Suzuki A, Suzuki Y, Sugano S. Expression of miR‑26a exhibits a negative correlation with HMGA1 and regulates cancer progression by targeting HMGA1 in lung adenocarcinoma cells. Mol Med Rep. 2017;15(2):534–542. doi:10.3892/mmr.2016.6053
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.