Back to Journals » OncoTargets and Therapy » Volume 7
HCA519/TPX2: a potential T-cell tumor-associated antigen for human hepatocellular carcinoma
Authors Aref A, Hoa N, Ge L, Agrawal A, Dacosta-Iyer M, Lambrecht N, Ouyang Y, Cornforth A, Jadus M
Received 28 January 2014
Accepted for publication 28 February 2014
Published 13 June 2014 Volume 2014:7 Pages 1061—1070
DOI https://doi.org/10.2147/OTT.S61442
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Video abstract presented by Ahmed M Aref.
Views: 156
Ahmed M Aref,1–3 Neil T Hoa,3 Lisheng Ge,3 Anshu Agrawal,4 Maria Dacosta-Iyer,5,6 Nils Lambrecht,5,6 Yi Ouyang,5,6 Andrew N Cornforth,7 Martin R Jadus5,6,8
1Biological Science Department, Modern Sciences and Arts University, Faculty of Dentistry, Cairo, Egypt; 2Southern California Institute for Research and Education, Veterans Affairs Medical Center, Long Beach, CA, USA; 3Research Health Care Group, Veterans Affairs Medical Center Long Beach, CA, USA; 4Department of Medicine, Division of Basic and Clinical Immunology, University of California, Irvine, CA, USA; 5Pathology and Laboratory Medicine Department, Veterans Affairs Medical Center Long Beach, CA, USA; 6Department of Pathology and Laboratory Medicine, University of California, Irvine, CA, USA; 7California Stem Cells, Inc., CA, USA; 8Neuro-Oncology Program, Chao Comprehensive Cancer Center, University of California, Irvine, CA, USA
Background: Immunotherapy for human hepatocellular cancer (HCC) is slowly making progress towards treating these fatal cancers. The identification of new antigens can improve this approach. We describe a possible new antigen, hepatocellular carcinoma-associated antigen-519/targeting protein for Xklp-2 (HCA519/TPX2), for HCC that might be beneficial for T-cell specific HCC immunotherapy.
Methods: HCC was studied for the expression for 15 tumor-associated antigens considered useful for immunotherapy within three HCC cell lines (HepG2, Hep3B, and PLC/PRF/5), lymphocytes, non-cancerous livers, and clinical HCC. The expression of tumor antigenic precursor proteins (TAPPs) messenger RNA was first screened by reverse transcriptase quantitative real-time polymerase chain reaction.
Results: Four antigens (alpha fetoprotein, aspartyl/asparaginyl β-hydroxylase, glypican-3 and HCA519/TPX2) proved to be the best expressed TAPPs within the HCC specimens by molecular analyses. HCA519/TPX2 was detected by intracellular cell flow cytometry within HCC cell lines by using a specific antibody towards this TAPP. This antibody also detected the protein within primary HCCs. We synthesized two HCA519/TPX2 peptides (HCA519464–472 and HCA519351–359) which can bind to human leukocyte antigen (HLA)-A*0201. Dendritic cells pulsed with these peptides stimulated cytolytic T lymphocytes (CTLs). These killer T-cells lysed HLA-A*0201+ T2 cells exogenously loaded with the correct specific peptide. The CTLs killed HepG2 (HLA-A2+ and HCA519+), but not the Hep3B and PLC/PRF/5 cell lines, which are HCA519+ but HLA-A2-negative. In silico analysis reveals that HCA519/TPX2 has the inherent ability to bind to a very wide variety of HLA antigens.
Conclusion: HCA519/TPX2 is a viable immunotarget that should be further investigated within HCC patients.
Keywords: tumor immunity, cytolytic T-cells, HLA-A2, HCA519/TPX2
Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers throughout the world; its increasing prevalence is due to chronic infections with hepatitis B or C viruses.1 Treatment of established HCC with standard oncological therapies remains ineffective, and the average survival time is just several months.2 New therapies are still desperately needed, especially those that focus on earlier biological/causative processes. Immunotherapy may be useful for future adjunct treatments of HCC.
HCC takes many years, if not decades, to appear. This time provides ample opportunity to prevent the appearance of this cancer. Immunizing children with the recombinant Hepatitis (HepB) virus vaccine has significantly reduced the incidence of HCC in those vaccinated people in Taiwan.3 Although this HepB immunizing approach targets one of the HCC inducing viruses, there are no such options for Hepatitis C (HepC) infections. Overall, this suggests that HCC can be successfully abrogated, if patients are properly vaccinated and this prophylactic immunity prevents the initial tumor cells from establishing the cancer. Previous immunotherapeutic attempts to treat well-established HCC have not been as successful,4,5 perhaps because HCC utilizes a variety of immunosuppressive mechanisms that prevent effective anti-tumor immunity. The best time to vaccinate people is before this cancer becomes established. Immunoprevention was proposed to be more effective at a clinical level by inhibiting the establishment of the initial HCC clones by using HCC-specific antigens before HCC develops.6 A recent study applying the concept of immunoprevention to adenomatous polyps to forestall colon cancer concluded the sooner the vaccination began the more efficient it became.7 Vaccinations of chronically infected HepB or HepC individuals may present a great opportunity to prevent HCC, provided the right HCC-associated antigens are used.
One of the best-known antigens for HCC is alpha fetoprotein (AFP).8 AFP is not universally expressed in all liver cancers.9,10 AFP is expressed during fetal development and may tolerize the immune system and can impede various immunotherapies.4,5
Aspartyl/asparaginyl β-hydroxylase (ASPH),11,12 glypican-3 (GPC3)13 and hepatocellular carcinoma-associated antigen-587 (HCA587)14,15 are being investigated as possible targets for HCC. HCC is reported to express several common antigens found on many types of cancer; the alternative form of macrophage colony stimulating factor (altM-CSF),16 B cyclin,17 carcinoembryonic antigen (CEA),9 N-acetylglucosaminyltransferase V (GnT-V),18 melanoma antigen (MAGE),19,20 multidrug resistance protein-3 (MRP3),21 New York-Esophageal Squamous cell carcinoma-1 (NY-ESO-1),22 telomerase reverse transcriptase (tert),23 sarcoma, synovial, X-chromosome-related-2/synovial sarcoma X breakpoint 2 (SSX2),19 survivin,24 and Wilm’s tumor antigen-1 (Wt-1).25 One HCC-associated antigen previously defined by a humoral response is HCA519.14 HCA519, also known as targeting protein for Xklp-2 (TPX2), is a microtubule associated protein needed for HCC cell division. In this paper using HCC cell lines and clinical samples, we conclude that HCA519/TPX2 is highly found within all HCC tested (n=16) and the protein expressions within the HCC cells are comparable to that exhibited by AFP, ASPH and GPC3. Peptides derived from HCA519/TPX2 can be recognized by human CTLs via an human leukocyte antigen (HLA)-A*0201 restricted manner. So HCA519/TPX2 should be further investigated as a good target for HCC immunotherapy.
Materials and methods
Cell lines and cell culture
HepG2, Hep3B and PLC/PRF/5 cells were obtained from the American Type Culture Collection (Manassas, VA, USA). Peripheral-blood mononuclear cells (PBMC) from healthy normal HLA-A*0201+ donor were obtained after informed consent was signed at University of California, Irvine to generate the dendritic cells (DC) and CTLs. Frozen HLA-A2+ PBMC was collected from cancer patients undergoing various therapies at the University of Southern California (USC) Norris Comprehensive Cancer Center (Los Angeles, CA, USA) in the early 2000s. These pre-HIPAA collected samples were supplied to us by Dr Jeffrey Weber (USC).
Clinical HCC and liver tissue
This project was approved by the Veteran Affairs Medical Center, Long Beach Institutional Review Board before this project was initiated. We collected autopsy material not needed for diagnostic purposes. Next of kin signed the autopsy consent forms which allow scientific investigations to be performed with excess material not needed for pathological analysis. Ten autopsy samples were collected at our facility and the University of California, Irvine, Pathology Department (Irvine, CA, USA). The US Cooperative Human Tissue Network (CHTN, Vanderbilt University, Nashville, TN, USA) supplied us with nine other cases. Two patients were HepB-positive, while three were positive for HepC within our cohort.
Quantitative real-time polymerase chain reaction analysis
Reverse transcriptase real-time polymerase chain reaction (qRT-PCR) was done as previously described.26,27 Additional primers for AFP, ASPH, GPC3 and HCA519 were:
- AFP forward: 5′-AGCCACTTGTTGCCAACTCA-3′;
- Reverse: 5′-GCCAACACCAGGGTTTACTG-3′;
- ASPH forward: 5′ AGGAACAAGCAGTATATGAACCTC-3′;
- Reverse: 5′-TCCTGCTGTTCTTCCACAGG-3′
- GPC3 forward: 5′-ACCAAGTCCGCTCCTTCTTC-3′;
- Reverse: 5′-GCAAATCTGATCCTGGCACG-3′;
- HCA519 forward: 5′-TTGCTCTGGCTGGAATAGGG-3′;
Reverse: 5′-TCTGTGCGGAAGTGGAAGTC-3′.
Data is presented as the mean ΔCt score ± standard error of the means. Individual groups were analyzed to be significantly different from each other using Student’s t-tests; P<0.05 was considered statistically significant.
Antibodies
Antibodies against AFP, ASPH, GPC3, and TPX2/HCA519 were obtained from Santa Cruz Biotech Inc., (Dallas, TX, USA). Cells were stained by intracellular staining as previously described27,28 using the antibodies against the HCC antigens listed. Ten thousand cells were analyzed with a Becton-Dickinson Biosciences FACSCalibur™ flow cytometer (San Jose, CA, USA). Kolmogorov–Smirnov statistics tests showed significant differences in flow cytometry (P<0.05) between the experimental and isotypic controls.
Immunofluorescent staining of human HCC specimens
Archival HCC paraffin blocks (collected pre-HIPPA) were immunostained using the anti-HCA519/TPX2 antibody. The fluorescence images were captured using the Nikon Eclipse E600 fluorescence microscope system (Nikon Instruments, Melville, NY, USA) with the RTKE Spot Camera and Spot software (Diagnostic Instruments, Inc., Sterling Heights, MI, USA ). The levels of fluorescence expression were calculated by a set of parameters (area, integrated density, and mean gray value) from ImageJ software (National Institutes of Health, Bethesda, ML, USA) and expressed in average pixel intensity (API). Values were calculated using the formula (API = average integrated density – area of selected image mean fluorescence of background readings). Three randomly selected areas (100×100 pixels) per image were used to compare the fluorescence intensities between the various tissue samples.
Peptides
The sequence for HCA519/TPX2 was analyzed by SYFPEITHI (http://www.syfpeithi.de/) and BIMAS (http://www-bimas.cit.nih.gov/molbio/hla_bind/). These algorithms predict the strength of the peptide-major histocompatibility binding affinity. Peptide one, HCA519/TPX2464–472: KILEDVVGV, had an SYPFEITHI score for HLA-A*0201as 30 and BIMAS predicted this peptide had a 1,167-minute half-life dissociation time. Peptide two, HCA519/TPX2351–359: NLLPSKSSV was scored by SYFPEITHI as 26, while BIMAS predicted a 257 minute half-life time. Both peptides were synthesized by GenScript Corporation (Piscataway, NJ, USA). The influenza M158-66 (GILGFVFTL) peptide was previously made by Proimmune (Oxford, UK)26,27 served as the negative control.
Generation of cytotoxic T lymphocytes
PBMC from a HLA-A2+ normal donor was collected and then used to generate CTLs as previously described.26–28 Every 2 weeks the CTLs were restimulated with the Miltenyi Biotec (Bergisch Gladback, Germany) T-cell expansion beads with interleukins -2, -7 and -15 (PeproTech, Rocky Hill, NJ, USA). Cytotoxicity was done as reported earlier.26–28 Cytolytic values were considered significantly different if P<0.05 was achieved using the Student’s t-test.
Results
HCC failed to express many of the “universal” tumor antigens
Initial studies used three human HCC cell lines, HepG2, Hep3B and PLC/PRF/5, and a limited number of HCC autopsy samples. Total RNA was extracted from those specimens, and from peripheral blood lymphocytes and non-HCC containing livers. The last two groups provided the negative controls. Complementary (cDNA) was synthesized from those samples by using reverse transcriptase. qRT-PCR was performed using primers that detect various tumor antigen precursor proteins (TAPPs). 18S ribosomal RNA served as the internal control as we previously used.26,27
We first examined universal tumor antigens found on many cancer types such as liver, brain, lung, kidney, skin and other cancers.29,30 Such antigens included altM-CSF, B-cyclin, CEA, GnT-V, MRP3, NY-Eso-1, survivin, SSX2, tert, and Wt-1. Figure 1 summarizes this data obtained from the experiments done with nine of these common tumor antigens (survivin, Wt-1, B-cyclin, CEA, MRP3, altM-CSF, SSX2, tert, and GnT-V). The survivin, Wt-1, and SSX2 mRNAs were expressed at very low levels (higher ΔCt values are the result of having to undergo more PCR cycles until the threshold was reached). The CEA and MRP3 mRNA levels were equivalent to those displayed by the non-cancerous liver or peripheral blood derived lymphocytes, which should be devoid of tumor-associated antigens. The only universal antigen that appeared significantly expressed in the HCC was tert. Interestingly, tert was not highly expressed within the HCC cell lines. Thus, HCC samples showed little expression of the common universal antigens.
  | Figure 1 qRT-PCR profiles of human TAPPs from human PBMC, HCC cell lines, non-cancerous liver and HCC tissue. |
HCC does express many of the HCC-specific antigens
A more restricted HCC tumor antigen profile (AFP, ASPH, GPC3, HCA519/TPX2, HCA587) was investigated (Figure 2). With the exception of HCA587, higher antigen expression patterns were observed within the HCC compared to the non-HCC samples. These four TAPP mRNAs were elevated within the three HCC cell lines, with ΔCt scores between 11–19. These TAPPs had very low levels of mRNAs found within the human lymphocytes with ΔCt scores greater than 23.
AFP showed the most variable phenotype of these four TAPPs examined. The data for only four autopsy samples tested are shown in the panel from the 12 total HCC samples tested. Eight of these HCC displayed a ΔCt score of >24, indicative of very little mRNA. All three HCC cell lines displayed very high levels of the AFP transcripts. ASPH was detected within the lymphocytes and within the non-cancerous liver with ΔCt scores slightly below 20, suggesting the TAPP mRNA is moderately present within the normal tissue. The amount of ASPH mRNA coming from the HCC was lower than the values displayed by the HCC cell lines, since the cell lines are pure tumor, whereas, the clinical samples contain non-tumorous stromal elements.
The highest expressed TAPPs found within the HCC were GPC3 and HCA519. The amount of these antigens within the lymphocytes and normal livers was very low; the amount of mRNA coming from the HCC was elevated (P<0.05) when compared to the non-cancerous liver. Likewise, there was significantly more mRNA isolated from the HCC cell lines, when matched to the control lymphocytes.
HCA519/TPX2 is not detected within normal tissue
Tissue was obtained from a patient who died from a cardiovascular event. The tissues sampled included: heart, intestine, kidney, liver, lung, skeletal muscle, skin, and spleen. The total RNA was then examined for HCA519/TPX2 mRNA by qRT-PCR. The spleen showed a slight amount of HCA519/TPX2 with a ΔCt score of 21.1±0.0. All other tissues had scores >22.9, which were deemed negligible if any mRNA. Thus high HCA519/TPX2 mRNA expression only appears within HCC tissue.
HCA519/TPX2 is detected by intracellular flow cytometry within the HCC cell lines
We used intracellular flow cytometry to quantitatively measure the amount of this HCA519/TPX2 TAPP present to confirm the molecular results. Figure 3 showed the three HCC cells were significantly (P<0.05) more positive for HCA519/TPX2 than the control lymphocytes. These studies verified the previous qRT-PCR data within the clinical HCC. The expression of HCA519/TPX2 shows that comparable levels of the AFP, ASPH and GPC3 proteins were detected within the HCC cell lines by using their respective antibodies. A slight bimodal profile was observed with the PBMC with all antigens. We attribute this profile to the primary or secondary antibodies binding to the Fc receptors found on the monocytes or lymphocytes present in the PBMC.
HCA519/TPX2 protein is detected within HCC
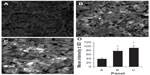
The expression of the HCA519/TPX2 protein is less well known than that of AFP, ASPH and GPC3. Two randomly chosen archival samples of HCC were examined by immunofluorescence staining using the same anti-HCA519/TPX2 antibody that was just used for intracellular flow cytometry. Minimal staining of the non-cancerous liver tissue is displayed in Figure 4A. The two HCC cancers were positively stained (Figure 4B and C) with the anti-HCA519/TPX2 antibody. Three representative fields of each sample were digitally quantitated and are presented in Figure 4D.
HLA-A2 restricted cytolytic T-cell responses are induced by HCA519/TPX2
We analyzed the human HCA519/TPX2 protein sequence by two epitope predicting algorithms (SYFPEITHI and BIMAS) and found two peptides that could have representative affinities for HLA-A*0201. DC from a normal HLA-A0201+ individual was pulsed with one of the two HCA519/TPX2 peptides overnight. The mature DC was incubated with autologous naïve CD8 T-cells. Once sufficient CTLs were expanded, they were used in standard 51Cr release assays.
The CTLs were first tested against human HLA-A*0201+ T2 hybridoma cells which can be loaded with exogenous peptides26,27 (Figure 5A and B). The T2 cells were loaded with either HCA519 peptide-1, peptide-2, or with the influenza M1 peptide. The HCA519464–472–specific CTLs (peptide 1) only killed the T2 cells exogenously loaded with the correct HCA519464–472 (peptide 1). Peptide 1 CTLs didn’t respond to T2 cells loaded with the wrong peptides (Figure 5A). Likewise, the HCA519351–359 CTL (peptide 2) specifically killed the T2 cells loaded with the appropriate HCA519351–359 peptide (Figure 5B). Thus, these CTLs proved specific for their correct antigens.
  | Figure 5 Human CTLs specifically kill target cells displaying HCA peptides via an HLA-A*0201 restricted manner. |
The CTLs were next tested against HepG2, Hep3B, and PLC/PRF/5 HCC cell lines. All three HCC cell lines were HCA519+, but only HepG2 is HLA-A2+.8 Both sets of the HCA519 specific CTLs killed the HepG2 cells (Figure 6), but failed to lyse the Hep3B or PLC/PRF/5 cells. Thus, HepG2 cells process sufficient HCA519 peptides to adequately allow the CTLs to recognize them in the context of HLA-A*0201.
Discussion
Cell-mediated immune responses, particularly those induced by human DC pulsed with tumor-derived material (killed tumor cells or peptides), are generating good clinical responses for several lethal tumor types.31,32 Therapeutic immune-based treatments are further enhanced against a variety of cancers, notably those that break the immunosuppressive pathways imposed upon the effector lymphocytes.33,34 DC-based vaccines have been used for HCC34,35 with little success, although these early studies were not combined with the newer methods to inhibit the so-called “check-point inhibitory antibodies”; ie, anti-CTLA-4, anti-PD-1, or anti-PD-L1 antibodies. These check-point inhibitory pathways are generating durable clinical responses within some patients even with moderate cancer burdens. For immunotherapies to work, the correct tumor antigens need to be recognized by the T-cells while under the right environmental conditions, so that the immune system can successfully combat the tumor. Finding more tumor-specific antigens is therefore paramount to the success of this modality. In this report, we believe that HCA519/TPX2 might be a promising tumor antigen for HCC T-cell-based immunotherapy.
We initially studied common universal tumor antigens that are widely reported to be found on many tumor types. We failed to find any expression of these antigens on either HCC cell lines and within clinical HCC samples (Figure 1). HCC-specific antigens, AFP, ASPH, GPC3, HCA519 and HCA587 were next examined by qRT-PCR (Figure 2). Barring HCA587, we found that the HCC material expressed higher mRNA levels (lower ΔCt scores) than either non-HCC liver tissue or lymphocytes. AFP specific transcripts were seen in four of the 12 samples tested, which is consistent with other studies that show AFP is quite variable within HCC patients.9,10 ASPH mRNA was highly displayed within HCC specimens, but unexpectedly high amounts of transcripts were also detected within the lymphocytes and non-cancerous liver. ASPH protein was not detected within the lymphocytes, even though they contain some mRNA (Figure 3). Otherwise the intracellular flow cytometry confirmed the results of the qRT-PCR when validated with the HCC cell lines.
HCA519/TPX2 was significantly elevated (P<0.05) within human HCC autopsy samples when compared to the livers derived from non-HCC containing livers and normal lymphocytes (Figure 2). HCA519 protein was detected by immunofluorescence within two randomly chosen HCC autopsy paraffin blocks from our archive (Figure 4). Using peptide/major histocompatibility antigen predicting databases, we chose two peptides to evaluate for possible T-cell epitopes that could be immunologically useful for this TAPP. Peptides HCA519464–472 and HCA519351–359 proved effective in generating HLA-A*0201 restricted CTLs. These CTLs only killed those T2 hybridoma cells that were exogenously loaded with their correct peptide (Figure 5). These CTLs also lysed the HepG2 cells which are both HCA519+ and HLA-A0201+ (Figure 6). However, the CTLs didn’t kill the HCA519+ Hep3B or PLC/PRF/5 cells which are HLA-A2 negative. This work suggests that HCC cell lines properly process sufficient HCA519 epitopes, so that CTLs can recognize and kill HCC cells with the proper HLA-A2 restriction.
Half of the US general population is HLA-A2+ and there were 118 other possible epitopes that might induce human immune responses using nonamers and decamers of the HCA519 TAPP with SYFPEITHI scores of 16 or greater. Further SYFPEITHI analysis of HCA519/TPX2 revealed there are numerous other epitopes for the other HLA phenotypes. Equivalent numbers of potential epitopes are predicted for HLA-A11 (104), HLA-A26 (102), and HLA-A68 (81). HLA-A3 had the most putative nonomer and decamer peptides (187). The only haplotype that had few binding peptides (28) was HLA-A24, so for Oriental and Western Pacific populations. HCA519/TPX2 might not be as effective as a target. When we examined potential peptides predicted to bind to HLA-B and HLA-DRB1 (MHC class II), there were many possible epitopes (>100) available for immune recognition by both CD8+ and CD4+ T-cells, respectively. So even though HCA519 might not be useful for HLA-A24+ individuals per se, the HCA519/TPX2 TAPP displays a high “epitope promiscuity” making this a possible universal vaccine for HCC. HCA519/TPX2 is therefore a potentially immunogenic molecule that can be applicable for most people across the planet. An alternative possibility is to use this antigen to vaccinate patients who have a higher predilection for HCC, such as those who have been infected by HepB or HepC and display chronic infections. Since HCC takes many years to arise, there is an adequate opportunity to develop immunity before the cancer establishes itself, especially if this antigen targets a molecule that plays a major role in HCC cell division. As the lessons learned from the patients vaccinated when human adenomatous polyps were detected, the sooner the vaccination began the more effective it became to prevent the colon cancer.7
HCA519/TPX2 associates with microtubules via kinetochores and facilitates aurora kinase A binding to the microtubules.36 Enhanced TPX2 production has also been associated with pancreatic37 and lung cancers.38 Studies by Tanaka et al39 and Yang et al40 show upregulation of TPX2 within HCC by microarray analysis. Satow et al41 demonstrated higher expression of HCA519/TPX2 in HCC tumor tissue compared to adjacent non-tumor tissue. Small interfering (siRNA)-mediated knockdown of this gene inhibited the proliferation of HCC cells and prevented the growth of HCC xenografts injected into immunodeficient mice. So this antigen might be at a key spot in the HCC replication cycle, where immunotherapy could have an immediate effect. Even if only the highly expressing HCA519/TPX2+ HCC cells are killed by activated CTLs, then the HCC tumor cell population as a whole might be sufficiently reduced, thereby hindering the in vivo tumor growth and in turn providing the patient with a longer survival. Thus, HCA519/TPX2 may be developed into future immunotherapy for HCC, which targets a key protein that these cancer cells need for their proliferation.
Acknowledgment
Funded in part by a Veterans Affairs Merit Review Grant.
Disclosure
The authors declare no conflicts of interest in this work.
References
Michielsen PP, Francque SM, van Dongen JL. Viral hepatitis and hepatocellular carcinoma. World J Surg Oncol. 2005;3:27. | |
Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27(9):1485–1491. | |
Chang MH, Chen TH, Hsu HM, et al. Prevention of hepatocellular carcinoma by universal vaccination against hepatitis B virus: the effect and problems. Clin Cancer Res. 2005;11(21):7953–7957. | |
Pardee AD, Butterfield LH. Immunotherapy of hepatocellular carcinoma: Unique challenges and clinical opportunities. Oncoimmunology. 2012;1(1):48–55. | |
Buonaguro L, Petrizzo A, Tagliamonte M, Tornesello ML, Buonaguro FM. Challenges in cancer vaccine development for hepatocellular carcinoma. J Hepatol. 2013;59(4):897–903. | |
Forni G, Lollini PL, Musiani P, Colombo MP. Immunoprevention of cancer: is the time ripe? Cancer Res. 2000;60(10):2571–2575. | |
Kimura T, McKolanis JR, Dzubinski LA, et al. MUC1 vaccine for individuals with advanced adenoma of the colon: a cancer immunoprevention feasibility study. Cancer Prev Res (Phila). 2013;6(1):18–26. | |
Vollmer CM Jr, Eilber FC, Butterfield LH, et al. Alpha-fetoprotein-specific genetic immunotherapy for hepatocellular carcinoma. Cancer Res. 1999;59(13):3064–3067. | |
Chu PG, Ishizawa S, Wu E, Weiss LM. Hepatocyte antigen as a marker of hepatocellular carcinoma: an immunohistochemical comparison to carcinoembryonic antigen, CD10, and alpha-fetoprotein. Am J Surg Pathol. 2002;26(8):978–988. | |
Fujioka M, Nakashima Y, Nakashima O, Kojiro M. Immunohistologic study on the expressions of alpha-fetoprotein and protein induced by vitamin K absence or antagonist II in surgically resected small hepatocellular carcinoma. Hepatology. 2001;34(6):1128–1134. | |
Lavaissiere L, Jia S, Nishiyama M, et al. Overexpression of human aspartyl(asparaginyl)beta-hydroxylase in hepatocellular carcinoma and cholangiocarcinoma. J Clin Invest. 1996;98(6):1313–1323. | |
Shimoda M, Tomimaru Y, Charpentier KP, Safran H, Carlson RI, Wands J. Tumor progression-related transmembrane protein aspartate-beta-hydroxylase is a target for immunotherapy of hepatocellular carcinoma. J Hepatol. 2012;56(5):1129–1135. | |
Komori H, Nakatsura T, Senju S, et al. Identification of HLA-A2- or HLA-A24-restricted CTL epitopes possibly useful for glypican-3-specific immunotherapy of hepatocellular carcinoma. Clin Cancer Res. 2006;12(9):2689–2697. | |
Wang Y, Han KJ, Pang XW, et al. Large scale identification of human hepatocellular carcinoma-associated antigens by autoantibodies. J Immunol. 2002;169(2):1102–1109. | |
Li B, Wang Y, Chen J, Wu H, Chen W. Identification of a new HLA-A*0201-restricted CD8+ T cell epitope from hepatocellular carcinoma-associated antigen HCA587. Clin Exp Immunol. 2005;140(2):310–319. | |
Ge L, Zhang JG, Samathanam CA, et al. Cytotoxic T cell immunity against the non-immunogenic, murine, hepatocellular carcinoma Hepa1-6 is directed towards the novel alternative form of macrophage colony stimulating factor. Cell Immunol. 2009;259(2):117–127. | |
Covini G, Chan EK, Nishioka M, Morshed SA, Reed SI, Tan EM. Immune response to cyclin B1 in hepatocellular carcinoma. Hepatology. 1997;25(1):75–80. | |
Shao DM, Wang QH, Chen C, et al. N-acetylglucosaminyltransferase V activity in metastatic models of human hepatocellular carcinoma in nude mice. J Exp Clin Cancer Res. 1999;18(3):331–335. | |
Bricard G, Bouzourene H, Martinet O, et al. Naturally acquired MAGE-A10- and SSX-2-specific CD8+ T cell responses in patients with hepatocellular carcinoma. J Immunol. 2005;174(3):1709–1716. | |
Chen Z, Shao JB, Wu W. Expression of A, G and B melanoma antigen genes in human hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2002;1(4):570–573. | |
Nies AT, Konig J, Pfannschmidt M, Klar E, Hofmann WJ, Keppler D. Expression of the multidrug resistance proteins MRP2 and MRP3 in human hepatocellular carcinoma. Int J Cancer. 2001;94(4):492–499. | |
Luo G, Huang S, Xie X, et al. Expression of cancer-testis genes in human hepatocellular carcinomas. Cancer Immun. 2002;2:11. | |
Mizukoshi E, Nakamoto Y, Marukawa Y, et al. Cytotoxic T cell responses to human telomerase reverse transcriptase in patients with hepatocellular carcinoma. Hepatology. 2006;43(6):1284–1294. | |
Ikeguchi M, Ueda T, Sakatani T, Hirooka Y, Kaibara N. Expression of survivin messenger RNA correlates with poor prognosis in patients with hepatocellular carcinoma. Diagn Mol Pathol. 2002;11(1):33–40. | |
Perugorria MJ, Castillo J, Latasa MU, et al. Wilms’ tumor 1 gene expression in hepatocellular carcinoma promotes cell dedifferentiation and resistance to chemotherapy. Cancer Res. 2009;69(4):1358–1367. | |
Zhang JG, Eguchi J, Kruse CA, et al. Antigenic profiles of glioma cells to generate allogeneic vaccines or DC-based therapeutics. Clin Cancer Res. 2007;13:566–575. | |
Ge L, Cornforth AN, Hoa NT, et al. Differential glioma-associated tumor antigen expression profiles of human glioma cells grown in hypoxia. PLoS One. 2012;7(9):e42661. | |
Agrawal S, Gupta S, Agrawal A. Human dendritic cells activated via dectin-1 are efficient at priming Th17, cytotoxic CD8 T and B cell responses. PLoS One. 2010;5(10):e13418. | |
Gordan JD, Vonderheide RH. Universal tumor antigens as targets for immunotherapy. Cytotherapy. 2002;4(4):317–327. | |
Cheever MA, Allison JP, Ferris AS, et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res. 2009;15(17):5323–5337. | |
Gulley JL, Drake CG. Immunotherapy for prostate cancer: recent advances, lessons learned, and areas for further research. Clin Cancer Res. 2011;17(12):3884–3891. | |
Prins RM, Soto H, Konkankit V, et al. Gene expression profile correlates with T-cell infiltration and relative survival in glioblastoma patients vaccinated with dendritic cell immunotherapy. Clin Cancer Res. 2011;17(6):1603–1615. | |
Pardoll D, Drake C. Immunotherapy earns its spot in the ranks of cancer therapy. J Exp Med. 2012;209(2):201–209. | |
Couzin-Frankel J. Immune therapy steps up the attack. Science. 2010;330(6003):440–443. | |
Lee WC, Wang HC, Hung CF, Huang PF, Lia CR, Chen MF. Vaccination of advanced hepatocellular carcinoma patients with tumor lysate-pulsed dendritic cells: a clinical trial. J Immunother. 2005;28(5):496–504. | |
Bird AW, Hyman AA. Building a spindle of the correct length in human cells requires the interaction between TPX2 and Aurora A. J Cell Biol. 2008;182(2):289–300. | |
Warner SL, Stephens BJ, Nwokenkwo S, et al. Validation of TPX2 as a potential therapeutic target in pancreatic cancer cells. Clin Cancer Res. 2009;15(21):6519–6528. | |
Ma Y, Lin D, Sun W, et al. Expression of targeting protein for xklp2 associated with both malignant transformation of respiratory epithelium and progression of squamous cell lung cancer. Clin Cancer Res. 2006;12(4):1121–1127. | |
Tanaka S, Arii S, Yasen M, et al. Aurora kinase B is a predictive factor for the aggressive recurrence of hepatocellular carcinoma after curative hepatectomy. Br J Surg. 2008;95(5):611–619. | |
Yang CW, Su JY, Tsou AP, et al. Integrative genomics based identification of potential human hepatocarcinogenesis-associated cell cycle regulators: RHAMM as an example. Biochem Biophys Res Commun. 2005;330(2):489–497. | |
Satow R, Shitashige M, Kanai Y, et al. Combined functional genome survey of therapeutic targets for hepatocellular carcinoma. Clin Cancer Res. 2010;16(9):2518–2528. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.