Back to Journals » Cancer Management and Research » Volume 12
HAND2-AS1 Inhibits Gastric Adenocarcinoma Cells Proliferation and Aerobic Glycolysis via miRNAs Sponge
Authors Xu Z, Lv H, Wang Y, Hu C, Chen S, Du Y, Shi C, Cheng X
Received 11 July 2019
Accepted for publication 9 February 2020
Published 1 May 2020 Volume 2020:12 Pages 3053—3068
DOI https://doi.org/10.2147/CMAR.S222878
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Zhiyuan Xu,1,* Hang Lv,2,* Yiping Wang,2 Can Hu,3 Shangqi Chen,3 Yian Du,1 Chengwei Shi,3 Xiangdong Cheng1
1Department of Gastric Surgery, Institute of Cancer and Basic Medicine, Cancer Hospital of the University of Chinese Academy of Sciences, Zhejiang Cancer Hospital, Hangzhou, Zhejiang 310022, People’s Republic of China; 2Key Laboratory of Integrated Traditional Chinese and Western Medicine for Diagnosis and Treatment of Digestive System Tumor, Hangzhou 300020, Zhejiang, People’s Republic of China; 3Department of Gastrointestinal Surgery, The First Affiliated Hospital of Zhejiang Chinese Medical University, Hangzhou 310006, Zhejiang, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiangdong Cheng
Department of Gastric Surgery, Institute of Cancer and Basic Medicine, Cancer Hospital of the University of Chinese Academy of Sciences, Zhejiang Cancer Hospital, No. 1 Banshan Road, Hangzhou 310022, Zhejiang, People’s Republic of China
Tel/Fax +86-571-87070965
Email [email protected]
Objective: To study the effect of lncRNA HAND2-AS1 on gastric adenocarcinoma (GA) cell property and explore its specific mechanism.
Methods: Data on stomach adenocarcinoma (STAD) were analyzed to screen differentially expressed lncRNA HAND2-AS1. RNA22-HAS database and dual luciferase reporter assay were applied to confirm the target relationship between HAND2-AS1/HIF3A and miR-184. The HAND2-AS1 and miR-184 expressions in tissue or cells were determined by qRT-PCR and Western blot. Besides, after GA cells (AGS) cultured in normoxic and hypoxic condition, phosphoenolpyruvate (PEP) and lactic acid were quantified by Phosphoenolpyruvate Fluorometric Assay Kit and Lactic Acid Detection kit, respectively. Additionally, colony formation assay, transwell invasion and migration assays were used to evaluate the abilities of cell invasion, migration, and proliferation in distinct conditions.
Results: The HAND2-AS1 and HIF3A expressions were down-regulated and miR-184 expression was up-regulated in GA tissues and cells. Dual luciferase reporter assay confirmed HAND2-AS1 and HIF3A were targeted by miR-184. AGS cell proliferation abilities were restrained by HAND2-AS1 and HIF3A overexpression and enhanced by miR-184, as well as migration and invasion abilities. In addition, HAND2-AS1 rescued enhanced AGS cell proliferation, cell migration, cell invasion abilities and glycolytic process caused by hypoxia via miR-184/HIF3A.
Conclusion: LncRNA HAND2-AS1 could inhibit GA cell proliferation, migration and invasion abilities and glycolytic process induced by hypoxia through miR-184/HIF3A signaling.
Keywords: gastric adenocarcinoma, lncRNA, cell proliferation, hypoxia, lactic acid
Introduction
Gastric cancer (GC) was regarded as one of the most pervasive cancers in China and caused more than 300,000 deaths every year.1 Moreover, GC has become a major health burden worldwide. Due to the lack of early detection strategies, preliminary diagnosis of GC is usually difficult, and GC is often with a dismal outcome.2 It is noted that approximately 85% of GCs were gastric adenocarcinomas (GA). A complex, multistep process is involved in the growth of GC, containing various genetic and epigenetic alterations of oncogenes, anti-oncogenes, DNA repair genes, cell cycle regulators, and signaling molecules.2 Therefore, it is imperative to have an inquiry into the molecular mechanisms underlying GA.
The latest studies have indicated that a group of long (>200 bp) non-coding RNAs (lncRNAs) could play a crucial role in tumorigenesis and might be used as biomarkers in omnifarious cancers.2 Pervasively transcribed in the genome, LncRNAs exert limited protein coding latent capacity and are hoped to be new candidates in carcinogenesis due to their diverse gene regulation functions in epigenetic mechanisms, transcriptional, and posttranscriptional.3 These evidences suggested that aberrant expression of lncRNAs might be a substantial contributor during the development of cancer. Researchers have shown that in GA cells, long non-coding RNAs were correlated with cancer-related proliferation or metastasis, which including HOTAIR, ANRIL, MRUL, CCAT1, NEAT1 and HULC.1,4,5 Despite the above findings, the limited knowledge of the functional roles and expressional patterns of lncRNAs in GA was uncovered until now. Based on previous researches, we administrated a microarray analysis using data from The Cancer Genome Atlas (TCGA) database to screen out the discrepancy expressions of lncRNAs between GA tissues and adjacent tissues. HAND2-AS1, with its down-regulated expressing in GA tumor tissues, became our research object.
HAND2-AS1 is located on chromosome 4q33-34 in a head-to-head orientation with Heart and Neural Crest Derivatives Expressed 2 (HAND2) gene. Previous studies have revealed that HAND2-AS1 overexpression was associated with neuroblastoma at stage IVS.6 Although there have been several existed studies about lncRNA HAND2-AS1 or HAND2 gene, for example, HAND2-AS1 and HAND2 were found to be co-regulated by a bidirectional promoter in neuroblastoma,7 the role of HAND2-AS1 in GA remains unclear. Moreover, whether miRNAs participate in the process of HAND2-AS1 down-regulation in GA still needs further exploration.
Hypoxia is a condition of reduced oxygen concentration that is often present in solid tumors including GA.8 Besides, hypoxia is one of the common microenvironment features in solid tumors and is relevant to tumor invasiveness, radio-insensitivity and chemo-resistance, as well as increases metastatic potential and poor survival rate.9 The tumor cells respond to the hypoxic conditions by inducing genes that regulate various biological processes including cellular proliferation, apoptosis, metabolism, angiogenesis and migration.8 Hypoxia-inducible factors (HIFs) are transcription factors, which are commonly expressed in mammals, including humans.10 In humans, three distinct forms of HIF-A have been identified (HIF-1A, HIF-2A, and HIF-3A), of which HIF3A is one hypotype of the HIF family. HIF-1A, HIF-2A, and HIF-3A are involved in regulating transcriptional programs in response to hypoxia.11 Previous reports have suggested that HIF3A could be directly regulated by miR-429 in human endothelial cells.12 Also, overexpression of microRNA-210 promoted chondrocyte multiplication and extracellular matrix deposition by targeting HIF-3alpha in osteoarthritis.13 However, the correlation between lncRNA and HIF3A in GA has not been discovered.
MicroRNAs (miRNAs) are a category of small, noncoding single-strand RNAs that can regulate the expression of multiple protein-coding genes at the post-transcriptional level by combining with the 3ʹ-UTR (3ʹ-untranslated region) of their target mRNAs.14 MiRNA expression profiling in human cancers has revealed signatures that are closely related to diagnosis, staging, tumor progression and therapies.15,16 Recent studies also suggested that lncRNAs could act as decoys for microRNAs.17 For instance, MEG3, a long non-coding RNA, could sponge miR-770 and thus influenced the development of gastric cardia adenocarcinoma.3 BANCR, another non-coding RNA that activated by BRAF, promoted gastric cancer cells proliferation.18 Furthermore, several researches have demonstrated that miRNAs were targeting HIF3A to regulate tumor progression. For instance, miR-210-3p and miR-485-5p were found to regulate HIF3A in soft tissue sarcoma cells.8 Up-regulation of microRNA-210 promoted chondrocyte proliferation and extracellular matrix deposition by targeting HIF-3alpha in osteoarthritis.13 MiR-429 was involved in regulation of the HIF3A expression in human endothelial cells.12
In the present study, the exploration of regulation among lncRNA HAND2-AS1, miRNA, mRNA and hypoxia in GA was performed. We administrated various experiments including bioinformatic prediction and distinct molecular biological tests to investigate the expression pattern of a cancer-related lncRNA HAND2-AS1 in GA, determine the association of HAND2-AS1 with miR-184 and HIF3A in GA cells under hypoxic condition, as well as characterize their functional roles in GA cells.
Methods
Clinical Specimens
All patients enrolled in this study were diagnosed as GA by pathologic examination. And they were all not treated with chemoradiotherapy or other treatment for tumor pre-operation. Clinicopathologic information of enrolled patients was shown in Table 1. Isolated specimens were stored in −80°C until next detection. Written informed consent was obtained from each participant and this study was approved by the Research Ethics Committee of Zhejiang Cancer Hospital.
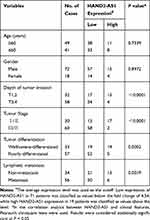 |
Table 1 Clinicopathologic Correlation of lncRNA HAND2-AS1 Expression in Human Gastric Adenocarcinoma |
Cell Lines and Culture Conditions
Human gastric adenocarcinoma AGS and NCI-N87 cell lines and human gastric mucosa GES 1 cell line were purchased from BeNa Culture Collection (http://www.bnbio.com). All of the cell lines were free of mycoplasma contamination (tested by the vendors using the MycoAlert kit from Lonza). No cell lines used in this study are found in the database of commonly misidentified cell lines (ICLAC and NCBI Biosample) based on short tandem repeats (STR) profiling performed by vendors. AGS and GES 1 cells were maintained in RPMI-1640 medium (Hyclone,South Logan,UT,USA) with 10% FBS. NCI-N87 cells were cultured in F-12 medium (Thermo Fisher Scientific,Waltham, MA,USA) with 10% FBS. Cells with 60-70% confluence were cultured in a routine incubator with the condition of 37°C, 5% CO2 or in a hypoxic incubator with the condition of 37°C, 5% CO2, 94% N2 and 1% O2.
Microarray Analysis
The data on STAD (Stomach Adenocarcinoma) were downloaded from TCGA. A total of 27 paired GA and adjacent tissue were included in current analysis to screen differentially expressed lncRNA. Raw data were normalized by DESeq2 library. Fold Change >2 and BH adjusted P < 0.05 and served as screening criteria. A series of analyses were performed by R programming language.
Cell Transfection
Recombinant plasmids HAND2-AS1-pcDNA3.1, miR-184 mimics, HIF3A-pcDNA3.1 and negative control were acquired from GenPharma pharmaceutical technology co. LTD. (Shanghai, China). Cells transfected with negative control, HAND2-AS1-pcDNA3.1, miR-184 mimics and the co-transfection of HAND2-AS1-pcDNA3.1 and miR-184 mimics were defined as NC, HAND2-AS1, miR-184 and HAND2-AS1+miR-184, respectively. Cell transfection was conducted using Lipofectamine 2000 (Invitrogen,Carlsbad,CA,USA) according to the instructions of manufacturer.
RNA Isolation and qRT-PCR
Total RNA was isolated through TRIzol reagent (Invitrogen). To quantify the miR-184 expression, TaqMan MicroRNA assays (Life Technologies) were performed. To quantify miRNA and mRNA expression, after quantified by NanoDrop 2000 (Thermo Fisher Scientific Inc, USA), 200 ng of total RNA was reversely transcribed into cDNA using aReverTra Ace qRT-PCR Kit (Toyobo, Japan). Following, the real-time PCR analysis was performed using SYBR Green I(10,000×) (Solarbio, Beijing, China). The relative miRNA and mRNA expression levels were calculated using the 2−ΔΔCT method. GAPDH were used as internal control for the quantification ofmRNA, respectively. Primer sequences for qRT-PCR are shown in Table 2. Each experiment was repeatedly performed three times.
 |
Table 2 Primer Sequences for qRT-PCR |
Dual Luciferase Reporter Assay
HAND2-AS1-wild type/mutant and HIF3A-wild type/mutant (both were synthesized by GeneChem Technology Co., Ltd, China) were cloned into the pGL3-control plasmid (Promega, Beijing, China), respectively, to construct the luciferase reporter vector. The processed pGL3-control plasmids, the pRL-TK plasmid (Promega) expressing renilla luciferase, miR-184 mimics and negative control were co-transfected into 293T cells using the Lipofectamine 2000 Reagent (Invitrogen). The cells were harvested after incubation for 24–48 hrs and detected the firefly and renilla luciferase activity using Dual Luciferase Reporter Assay Kit (Promega). Firefly luciferase activity was standardized to Renilla luciferase activity and the foldchange was calculated by the miRNA compared with NC.
Colony Formation Assay
Transfected cells with density of 1 × 103 per well were seeded in 12 well plates, afterwards, cultured in RPMI-1640 medium with 10% FBS at 37°C with 5% CO2. The culture was terminated when visible cells were incubated. After discarding the supernatant, the cells were washed with pre-cold PBS and soon fixed by 4% paraformaldehyde for 15 min. Then, they were colorized with crystal violet for 10–30 min before dried in the air. An inverted microscope (Nikon, Tokyo, Japan) was used to count colonies. The experiment had at least three repetitions.
Migration and Invasion Assays
The cell migration assay was performed using transwell chambers (8-µm pore, Corning, USA), which placed into 24 well plates. Medium with 10% FBS, as chemoattractant, was added in the lower chamber. And 2 ×105 cells were added on the upper insert. The insert was incubated for 24 hrs. Then the insert was fixed, stained and counted. Besides, for the cell invasion assay, transwell chambers membranes were coated with Matrigel (Corning), and the procedure of the cell invasion assay was similar to the cell migration assay.
Western Blot
Cells or tissues were treated with RIPA lysis buffer (Beyotime, China) to harvest proteins. The concentration of protein was determined using BCA protein assay kit (Thermo Fisher, Waltham, USA). Proteins were resolved by 10% SDS denatured polyacrylamide gel and then transferred onto a nitrocellulose membrane. The membranes were blocked in 5% non-fat milk and incubated with following primary antibodies: anti-HIF3α (Abcam, Cambridge, MA, USA, ab10134, 1:500), anti-HIF1α (Abcam, ab16066, 1:2000) or anti-GAPDH (Abcam, ab8245, 1:5000) overnight at 4°C. Then, the membranes were washed with TBST for 3 times and incubated with HRP-conjugated secondary antibody. The signals of immunoreactive bands were visualized by the enhanced chemiluminescent (ECL) substrate.
Induction of Hypoxia
Hypoxia was produced in a CO2/O2 incubator (Binder CB160 with O2 control, trigas model) and oxygen level was additionally supervised with an independent oxygen meter (PCS GOX100). In brief, cells were cultured in 10 cm dishes at 1% O2 for the time periods specified. Control cells were maintained in normoxic condition in the same condition and harvested at the specified time.
Measurement of Biochemical Indicators
Phosphoenolpyruvate level was quantified in growth medium using Phosphoenolpyruvate Fluorometric Assay Kit (Biovision, CA, USA) according to the manufacturer’s instructions. The quantification was performed by spectrophotometer (Thermo, Waltham, MA, USA) at Ex/Em = 535 nm/587 nm. Besides, Lactic Acid Detection kit (KeyGen, Nanjing, China) was used to detect lactic acid concentration. The assay was performed following the manufacturer’s instructions. The detection was performed using a spectrophotometer (Thermo) at 570 nm.
Statistical Analysis
Data were present as mean ± standard deviation (mean ± SD). R software version 3.2.1 and GraphPad Prism 6.0 were used for statistical analysis. Student’s t-test was applied to calculate statistically significant differences. Statistical evaluation was performed using Pearson correlation test to analyze the association between HAND2-AS1 and HIF3A expression. P value of less than 0.05 was considered statistically significant.
Results
LncRNA HAND2-AS1 Expression Was Down-Regulated in GA
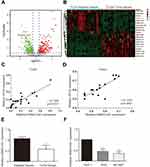
The data on STAD downloaded from TCGA were analyzed to screen differentially expression lncRNA with the criteria of Fold Change >2 and BH adjusted P < 0.05 (Figure 1A, P < 0.05). The twenty genes with the largest Fold Change value were selected to draw the heat map, of which lncRNA HAND2-AS1 expression was lower in GA tissues than that in adjacent tissues (Figure 1B). Scatter plot shows the positive correlation of HAND2-AS1 mRNA expression and HIF3A mRNA expression in TCGA and tissue sample (Figure 1C and D). The HAND2-AS1 expression was detected in 90 paired GA and adjacent tissues using qRT-PCR. The result showed that compared with adjacent tissues, the HAND2-AS1 expression was lower than that in the GA tissues (Figure 1E, P < 0.05). To further confirm the different expression of the HAND2-AS1 between GA and normal individuals, we determined the HAND2-AS1 expression in human gastric adenocarcinoma AGS and NCI-N87 cell lines and human gastric mucosa GES 1 cell line. The result suggested that the HAND2-AS1 expression was significantly lower both in AGS and NCI-N87 cells than that in GES 1 cell (Figure 1F, P < 0.05). Moreover, the difference between AGS cell and GES 1 cell was more obvious. Thus, AGS cell was applied for follow-up studies.
HAND2-AS1 Was Targeted by miR-184
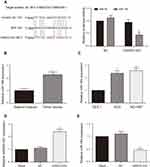
According to RNA22-HAS database, three binding sites targeted to miR-184 existed in 3ʹUTR of HAND2-AS1, positions 1384–1402, 4796–4816 and 4812–4830, respectively. There were only four complementary base pairings in the HAND2-AS1 3ʹUTR positions of 4796–4816. The binding sites in the HAND2-AS1 3ʹUTR positions of 1384–1402 and 4812–4830 are showed in Figure 2A. Dual luciferase reporter assay was used to verify the target relationship between HAND2-AS1 and miR-184. The results showed relative luciferase activity was significantly lower in the group of containing wild type HAND2-AS1 3ʹUTR sequence than that in the group of containing mutant HAND2-AS1 3ʹUTR sequence (Figure 2A, P < 0.05), indicating HAND2-AS1 was targeted by miR-184. Besides, we determined the miR-184 expression in GA and adjacent tissues, GA cells and gastric mucosa cell using qRT-PCR. Results showed that the miR-184 expression in GA tissues was higher than that in adjacent tissues (Figure 2B, P < 0.05). Compared with gastric mucosa GES 1 cell, the expression level of miR-148 was up-regulated both in the gastric adenocarcinoma AGS and NCI-N87 cells, of which more obvious up-regulation was observed in the AGS cell (Figure 2C, P < 0.05). In order to research the role of HAND2-AS1 in miR-184 expression, we successfully constructed the AGS cell with HAND2-AS1 up-regulation (Figure 2D, P < 0.05). Meanwhile, the miR-184 expression in the AGS cells with or without HAND2-AS1 up-regulation was detected by qRT-PCR. Results suggested the miR-184 expression was down-regulated after over-expressed HAND2-AS1 in the AGS cell (Figure 2E, P < 0.05). Above-mentioned results consistently indicated the target relationship between HAND2-AS1 and miR-184.
HAND2-AS1 and miR-184 Regulated AGS Cell Property
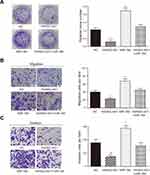
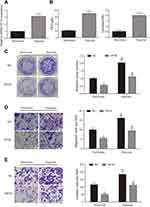
To study the effect of HAND2-AS1 and miR-184 on AGS cell property, we successfully constructed the AGS cells overexpressing HAND2-AS1, miR-184, both HAND2-AS1 and miR-184. Then, colony formation assay, transwell migration and invasion assays were used to detect transfected AGS cell property. In the colony formation assay, compared with normal AGS cells (NC), clone number was significantly down-upregulated in the AGS cells that overexpressing HAND2-AS1, but significantly up-regulated in the AGS cells that overexpressing miR-184. However, clone number in the AGS cells overexpressing both HAND2-AS1 and miR-184 was not different from that in NC cell (Figure 3A, P < 0.05). In transwell migration and invasion assays, results showed that in the AGS cells, cell migration and invasion ability was inhibited by HAND2-AS1, but enhanced by miR-184. Similarly, overexpressing both HAND2-AS1 and miR-184 did not notably change cell migration and invasion abilities in the AGS cells (Figure 3B and C, P < 0.05). In brief, cell proliferation, migration and invasion were decreased by HAND2-AS1 and increased by miR-184 in AGS cells. Overexpressing both HAND2-AS1 and miR-184 in the AGS cells relieved cell property alteration caused by HAND2-AS1 or miR-184, to some extent, confirming again that HAND2-AS1 was targeted by miR-184.
HIF3A Was Targeted by miR-184
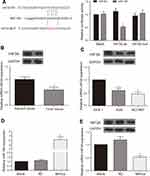
According to RNA22-HAS database, the HIF3A 3ʹUTR contained the predicted miR-184 binding site. The sequences of wild type and mutant HIF3A are shown in Figure 4A. Dual luciferase reporter assay was used to verify the target relationship between HIF3A and miR-184. The results showed that forced expression of miR-184 remarkably inhibited the luciferase activity in the group of containing wild type HIF3A 3ʹUTR, which suggested that HIF3A was the target of the miR-184 (Figure 4A, P < 0.05). Moreover, we determined the HIF3A expression in GA and adjacent tissues, GA cells and gastric mucosa cell using Western blot and qRT-PCR. Results showed that the HIF3A expression in GA tissues was lower than that in adjacent tissues (Figure 4B, P < 0.05). Compared with gastric mucosa GES 1 cell, the expression level of HIF3A was down-regulated both in the gastric adenocarcinoma AGS and NCI-N87 cells, of which more obvious down-regulation was observed in the AGS cell (Figure 4C, P < 0.05). To further investigate the role of miR-184 in HIF3A expression, we successfully constructed the AGS cell with miR-184 up-regulation (Figure 4D, P < 0.05). Simultaneously, Western blot and qRT-PCR were used to detect the HIF3A protein/mRNA expression in the AGS cells with or without miR-184 up-regulation. Results showed HIF3A expression was significantly down-regulated by miR-184 overexpression in the AGS cells (Figure 4E, P < 0.05). Those results revealed that HIF3A was targeted by miR-184.
HIF3A Inhibited AGS Cell Property Both in Normoxic and Hypoxic Environments
Based on the evidences that hypoxia enhances cancer cell property in many cancers and HIF3A is closely associated with hypoxia, we wanted to survey the effect of HIF3A on AGS cell property in normoxic and hypoxic conditions. Firstly, the detections of HIF1A, phosphoenolpyruvate (PEP) and lactic acid were performed by qRT-PCR, Phosphoenolpyruvate Fluorometric Assay Kit and Lactic Acid Detection kit, respectively, to verify the successful induction of hypoxia. Results showed the levels of HIF1A, PEP and lactic acid were significantly increased in the AGS cells with hypoxic induction, suggesting AGS cells were successfully induced by hypoxia (Figure 5A and B, P < 0.05). Secondly, AGS cells with or without overexpressing HIF3A were both cultured in normoxic or hypoxic condition to detect the effects of HIF3A and hypoxia on AGS cell property. Colony formation assay revealed hypoxia promoted AGS cell proliferation both in NC and HIF3A overexpression groups (Figure 5C, P < 0.05) and HIF3A inhibited AGS cell proliferation both in normoxic and hypoxic groups (Figure 5C, P < 0.05). Similarly, transwell migration and invasion assays indicated hypoxia promoted AGS cell migration and invasion both in NC and HIF3A overexpression groups (Figure 5D and E, P < 0.05) and HIF3A inhibited AGS cell migration and invasion both in normoxic and hypoxic groups (Figure 5D and E, P < 0.05).
HAND2-AS1 Regulated Glycolysis and Cell Property in AGS Cells Induced by Hypoxia
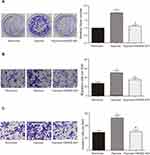
Anterior results suggested HAND2-AS1 was targeted by miR-184, HIF3A was targeted by miR-184 and HIF3A inhibited AGS cell proliferation, migration and invasion both in normoxic and hypoxic conditions. Thus, we wanted to investigate the regulation of HAND2-AS1 to AGS cells induced by hypoxia. The AGS cells cultured in normoxic or hypoxic condition and the AGS cells overexpressing HAND2-AS1 with hypoxia induction were chosen to perform follow-up studies. The results of qRT-PCR showed the HAND2-AS1 expression in AGS cells was lower in hypoxic condition than that in normoxic condition (Figure 6A, P < 0.05). Overexpressing HAND2-AS1 in AGS cells rescued the decrease of HAND2-AS1 caused by hypoxia (Figure 6A, P < 0.05), indicating AGS cells induced by hypoxia successfully overexpressed HAND2-AS1. The miR-184 expression was increased by hypoxia (Figure 6B, P < 0.05), and the increase caused by hypoxia was inhibited by the HAND2-AS1 overexpression (Figure 6B, P < 0.05). Moreover, Western blot and qRT-PCR revealed hypoxia increased HIF3A expression but decreased HIF1A expression, and HAND2-AS1 effectively relieved the alteration induced by hypoxia (Figure 6C and D, P < 0.05). Meanwhile, we studied the effect of HAND2-AS1 on the glycolysis in AGS cells. As shown in Figure 6E and F, high levels of PEP and lactic acid induced by hypoxia were suppressed by HAND2-AS1 in AGS cells (P < 0.05), suggesting HAND2-AS1 inhibited glycolysis process in AGS cells induced by hypoxia. In addition, cell proliferation, migration and invasion abilities of the cells in aforementioned three states were determined to research the effect of HAND2-AS1 on AGS cell property. Hypoxia promoted AGS cell proliferation, migration and invasion as expected (Figure 7A–C, P < 0.05). Nevertheless, HAND2-AS1 suppressed AGS cell proliferation, migration and invasion enhanced by hypoxia (Figure 7A–C, P < 0.05). Therefore, HAND2-AS1 could regulate glycolysis process and cell property in AGS cells induced by hypoxia.
Discussion
LncRNAs are emerging as the molecules that have a significant impact on many cellular processes. Moreover, lncRNAs have been proved to be associated with nearly every human cancer. Many lncRNAs demonstrate exquisite cellular-, tissue- or developmental stage-specific expression patterns.19 However, the associations between specific lncRNAs and the mechanistic role in driving GA and the function of lncRNAs in cancer cells under hypoxic condition have been unclear. In this study, LncRNA HAND2-AS1 expression was down-regulated in GA tissues and cells. Bioinformatic prediction found a novel signaling pathway of lncRNA HAND2-AS1/miR-184/HIF3A in GA. Then, dual luciferase report assay approved the target relationship between HAND2-AS1/HIF3A and miR-184. Besides, the expression of miR-184 was higher in GA tissues and cells than that in the adjacent tissues and gastric mucosa cells, and the expression of HIF3A was lower in GA tissues and cells than that in the adjacent tissues and gastric mucosa cells. GA cell invasion, migration and proliferation were suppressed by HAND2-AS1 and enhanced by miR-184. In addition, HAND2-AS1 could effectively relieve enhanced cell proliferation, migration and invasion induced by hypoxia in GA cells.
LncRNAs can exert distinct functions in diverse cancer. On one hand, most of the lncRNAs were reported to be highly expressed in various cancers, which played an accelerative role in oncogenesis. For instance, the study of Zhang et al demonstrated that up-regulation of lncRNA BANCR was observed both in gastric tumor cells and tissues, which also promoted cell growth and inhibited cell apoptosis in gastric cancer cells.18 Mizrahi et al reported that CCAT1 was highly expressed in GC cells and promoted the progression.5 Lee et al demonstrated that lncRNA HOTAIR inhibited apoptosis and promoted invasion, supporting that HOTAIR acted as promotor in progression of gastric cancer and carcinogenesis.20 On the other hand, several lncRNAs including lncRNA HAND2-AS1 as tumor suppressor involve in regulation of various cellular processes. Liu et al revealed that lncRNA loc285194 was down-regulated in colon tumor specimens and function as a p53-regulated tumor suppressor.21 Chunharojrith et al reported that expression of lncRNA MEG3 significantly suppressed xenograft tumor growth in nude mice and confirmed MEG3 as a tumor suppressor in the pituitary tumor.22 Similar to lncRNA loc285194 and lncRNA MEG3, a novel neoplasm suppressor, lncRNA HAND2-AS1 in GA was identified in this study. Results of current study indicated HAND2-AS1 in GA tissues and cells was lowly expressed, and overexpression of HAND2-AS1 suppressed cell proliferation, migration and invasion in GA cells, followed with the miR-184 upregulation in vitro.
Hypoxic stress induces the expression of an enlarging but specific subset of miRNAs.23 Experimental studies have demonstrated that hypoxia regulates miRNA expression and activity through several important mechanisms. Hypoxia regulates dynamic changes in hypoxamir maturation and function by processing complexes containing Drosha and Dicer.23 Experimental studies have demonstrated that hypoxia regulates miRNA expression and activity through several important mechanisms. Hypoxia regulates dynamic changes in hypoxamir maturation and function by processing complexes containing Drosha and Dicer.24 For example, exposure of rats to hypoxia for as little as 48 hrs restrained the expression of Dicer and Drosha mRNA and protein levels in the samples of lung tissue.25
These changes resulted in distinct and dynamic shifts in miRNA expression under hypoxia treatment. Hypoxia also mediates crucial post-translational modification (PTMs) that regulate the activity of proteins participated in the biogenesis and functionality of miRNAs.24 Recent study suggested that hypoxia promotes the expression of type I collagen prolyl-4-hydroxylase [C-P4H(I)], an enzyme that increases prolyl-hydroxylation and accumulation of Ago2, a vital component of the miRISC.26 Collectively, these data indicate that hypoxia modulates several phases of miRNA biogenesis, maturation, and function. Base on the results of the current study, we suspected that hypoxia regulates the facets of miR-184 transcription, maturation, and function which result in up-regulated in GC cells.
Increasing evidences demonstrated that lncRNAs involved in cellular processes through sponging the specific miRNA. In the present study, we found that lncRNA HAND2-AS1 regulated cell proliferation, migration and invasion in GA through targeting miR-184. In addition, the expression of miR-184 was up-regulated in GA and repressed by HAND2-AS1. However, Fang et al reported that lncRNA UCA1 promoted cell proliferation and enhanced cisplatin resistance of OSCC by suppressing miR-184.27 Similarly, Zhou et al revealed artesunate suppressed the viability and mobility of prostate cancer (PCa) cells through lncRNA UCA1, the sponge of miR-184.28 Consistent with aforementioned studies, the study from Emdad et al reported that suppression of miR-184 upregulated SND1 and promotes malignant gliomas aggressiveness.29 Oppositely, miR-184 turned up to be highly expressed and could enhance the malignant biological behavior via targeting FIH-1 in human glioma cell line A172.30 Moreover, miR-184 functions as an oncogenic regulator in hepatocellular carcinoma (HCC)31 and oral tongue squamous cell carcinoma (OTSCC).32 Therefore, definite mechanisms related to miR-184 in distinct cancer are differential and need to be severally elucidated.
Certain lncRNAs could bind and negatively influence miRNAs, which are substantially involved in the competing endogenous RNA (ceRNA) network, thereby regulating linear RNA transcription and protein production. Li et al reported that lncRNA PVT1-5 could strongly suppress miR-126 activity, resulting in increased levels of downstream target SLC7A5 and promoting cell proliferation of lung cancer.33 LncRNA H19 played a role in mechanical tension-induced osteogenesis of bone marrow mesenchymal stem cells by functioning as a sponge of miR-138.34 LncRNA MALAT1 as a competing endogenous RNA of miR-125b promoted sepsis-induced cardiac inflammation and dysfunction through modulating p38 MAPK/NFκB signaling pathway.35 In our study, target relationships were verified between HAND2-AS1 and miR-184 and between HIF3A and miR-184 and a novel signaling pathway of HAND2-AS1/miR-184/HIF3A in GA was identified. HIF‑3A is a target gene of HIF‑1, is closely related to hypoxia, and regulates the expression of hypoxic genes.10 Inducible HIF3A4, as a HIF3A splicing variant, impaired angiogenesis, proliferation and metabolism/oxidation in hypervascular malignant meningiomas.36 Considering the signaling pathway of HAND2-AS1/miR-184/HIF3A, therefore, we probed into the effect of HAND2-AS1 on GA cell under hypoxic condition. Finally, we found HAND2-AS1 could effectively inhibit GA cell proliferation, migration, and invasion in GA cell induced by hypoxia.
LncRNAs are either up or downregulated and modulate many key signaling pathways in response to hypoxia, which contribute to tumor hallmarks. The mechanism of hypoxia-induced lncRNAs expression partly due to hypoxia-inducing factor direct binding on the promoter region of lncRNAs. These lncRNAs exist oncogenic properties and facilitate cancer formation and progression. Moreover, lncRNAs modulate the HIF pathway by acting as antisense lncRNAs (aHIFα) or by functioning as microRNAs sponge (linc-ROR).37 In our case, HAND2-AS1 can form as microRNAs sponge (miR-184) in response to hypoxia which result the decrease expression of HAND2-AS1 in GC cells.
Inevitably, several limitations existed in this study. Whether other elements such as HIF1A participated in the inhibition of HAND2-AS1 to cell proliferation, migration and invasion in GA cell under hypoxic condition retains be further elucidated in the future. Additionally, direct experimental evidence demonstrating a functional link between hypoxia and the induction of miR-184 is currently lacking, and further work is needed to explain the relationship between hypoxia and miR-184. Furthermore, we found a tumor suppresser in GA cells under hypoxic condition, which could be expected to become a possible pre-diagnose target in the therapy of GA. In conclusion, HAND2-AS1 inhibited cell proliferation, migration and invasion of GA cells induced by hypoxia through miR-184/HIF3A signaling. HAND2-AS1 could be a novel latent target in GA.
Abbreviations
ceRNA, competing endogenous RNA; ECL, enhanced chemiluminescent; GA, gastric adenocarcinoma; GC, Gastric cancer; HAND2, Heart and Neural Crest Derivatives Expressed 2; HIFs, Hypoxia-inducible factors; HCC, hepatocellular carcinoma; HIFs, Hypoxia-inducible factors; lncRNAs, long (>200 bp) non-coding RNAs; miRNAs, MicroRNAs; NC, normal AGS cells; OTSCC, oral tongue squamous cell carcinoma; PEP, phosphoenolpyruvate; PCa, prostate cancer; PEP, phosphoenolpyruvate; STAD, stomach adenocarcinoma; TCGA, The Cancer Genome Atlas.
Data Sharing Statement
The dataset supporting the conclusions of this article is available in the corresponding author repository.
Ethics and Consent Statement
All procedures performed in studies involving human participants were in accordance with the ethical standards of Zhejiang Cancer Hospital. This article does not contain any studies with animals performed by any of the authors.
Informed Consent
Informed consent was obtained from all individual participants included in this study.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This study was supported by the Natural Science Foundation of Zhejiang Province (Grants no. LY18H290006), Zhejiang Provincial Medical and Healthy Science and Technology Projects (Grants No. WKJ-ZJ-1728, 2016KYB220), Program of Zhejiang Provincial TCM Sci-tech Plan (Grants no. 2016ZZ012, 2018ZY006, 2018ZB044), Zhejiang Provincial Science and Technology Projects (Grants no. 2018C37045), and National Natural Science Foundation of China (Grant No. 81573953, 81703753).
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Wang Z, Dai J, Hu N, et al. Identification of new susceptibility loci for gastric non-cardia adenocarcinoma: pooled results from two Chinese genome-wide association studies. Gut. 2017;66(4):581–587. doi:10.1136/gutjnl-2015-310612
2. Ma Y, Liu L, Yan F, et al. Enhanced expression of long non-coding RNA NEAT1 is associated with the progression of gastric adenocarcinomas. World J Surg Oncol. 2016;14(1):41. doi:10.1186/s12957-016-0799-3
3. Guo W, Dong Z, Liu S, et al. Promoter hypermethylation-mediated downregulation of miR-770 and its host gene MEG3, a long non-coding RNA, in the development of gastric cardia adenocarcinoma. Mol Carcinog. 2017;56(8):1924–1934. doi:10.1002/mc.v56.8
4. Song W, Liu YY, Peng JJ, et al. Identification of differentially expressed signatures of long non-coding RNAs associated with different metastatic potentials in gastric cancer. J Gastroenterol. 2016;51(2):119–129. doi:10.1007/s00535-015-1091-y
5. Mizrahi I, Mazeh H, Grinbaum R, et al. Colon Cancer Associated Transcript-1 (CCAT1) expression in adenocarcinoma of the stomach. J Cancer. 2015;6(2):105–110. doi:10.7150/jca.10568
6. Yang X, Wang CC, Lee WYW, et al. Long non-coding RNA HAND2-AS1 inhibits invasion and metastasis in endometrioid endometrial carcinoma through inactivating neuromedin U. Cancer Lett. 2018;413:23–34. doi:10.1016/j.canlet.2017.10.028
7. Voth H, Oberthuer A, Simon T, et al. Co-regulated expression of HAND2 and DEIN by a bidirectional promoter with asymmetrical activity in neuroblastoma. BMC Mol Biol. 2009;10(1):28. doi:10.1186/1471-2199-10-28
8. Gits CM, van Kuijk PF, de Rijck JC, et al. MicroRNA response to hypoxic stress in soft tissue sarcoma cells: microRNA mediated regulation of HIF3alpha. BMC Cancer. 2014;14(1):429. doi:10.1186/1471-2407-14-429
9. Winther M, Alsner J, Tramm T, et al. Prognostic value of hypoxia-regulated gene expression in loco-regional gastroesophageal cancer. Acta Oncol (Madr). 2016;55(5):652–655. doi:10.3109/0284186X.2015.1114680
10. Yang SL, Wu C, Xiong ZF, Fang X. Progress on hypoxia-inducible factor-3: its structure, gene regulation and biological function (Review). Mol Med Rep. 2015;12(2):2411–2416. doi:10.3892/mmr.2015.3689
11. Kai AK, Chan LK, Lo RC, et al. Down-regulation of TIMP2 by HIF-1alpha/miR-210/HIF-3alpha regulatory feedback circuit enhances cancer metastasis in hepatocellular carcinoma. Hepatology. 2016;64:473–487. doi:10.1002/hep.28577
12. Janaszak-jasiecka A, Bartoszewska S, Kochan K, et al. miR-429 regulates the transition between Hypoxia-Inducible Factor (HIF)1A and HIF3A expression in human endothelial cells. Sci Rep. 2016;6(1):22775. doi:10.1038/srep22775
13. Li Z, Meng D, Li G, et al. Overexpression of microRNA-210 promotes chondrocyte proliferation and extracellular matrix deposition by targeting HIF-3alpha in osteoarthritis. Mol Med Rep. 2016;13(3):2769–2776. doi:10.3892/mmr.2016.4878
14. Hu Y, Wang J, Qian J, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74(23):6890–6902. doi:10.1158/0008-5472.CAN-14-0686
15. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7(1):11215. doi:10.1038/ncomms11215
16. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
17. Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448(3):315–322. doi:10.1016/j.bbrc.2013.12.126
18. Zhang ZX, Liu ZQ, Jiang B, et al. BRAF activated non-coding RNA (BANCR) promoting gastric cancer cells proliferation via regulation of NF-kappaB1. Biochem Biophys Res Commun. 2015;465(2):225–231. doi:10.1016/j.bbrc.2015.07.158
19. Gibb EA, Warren RL, Wilson GW, et al. Activation of an endogenous retrovirus-associated long non-coding RNA in human adenocarcinoma. Genome Med. 2015;7(1):22. doi:10.1186/s13073-015-0142-6
20. Lee NK, Lee JH, Park CH, et al. Long non-coding RNA HOTAIR promotes carcinogenesis and invasion of gastric adenocarcinoma. Biochem Biophys Res Commun. 2014;451(2):171–178. doi:10.1016/j.bbrc.2014.07.067
21. Liu Q, Huang J, Zhou N, et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41(9):4976–4987. doi:10.1093/nar/gkt182
22. Chunharojrith P, Nakayama Y, Jiang X, et al. Tumor suppression by MEG3 lncRNA in a human pituitary tumor derived cell line. Mol Cell Endocrinol. 2015;416:27–35. doi:10.1016/j.mce.2015.08.018
23. Loscalzo J. The cellular response to hypoxia: tuning the system with microRNAs. J Clin Invest. 2010;120(11):3815–3817. doi:10.1172/JCI45105
24. Nallamshetty S, Chan SY, Loscalzo J. Hypoxia: a master regulator of microRNA biogenesis and activity. Free Radic Biol Med. 2013;64:20–30. doi:10.1016/j.freeradbiomed.2013.05.022
25. Caruso P, MacLean MR, Khanin R, et al. Dynamic changes in lung microRNA profiles during the development of pulmonary hypertension due to chronic hypoxia and monocrotaline. Arterioscler Thromb Vasc Biol. 2010;30(4):716–723. doi:10.1161/ATVBAHA.109.202028
26. Wu C, So J, Davis-Dusenbery BN, et al. Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of argonaute2. Mol Cell Biol. 2011;31(23):4760–4774. doi:10.1128/MCB.05776-11
27. Fang Z, Zhao J, Xie W, et al. LncRNA UCA1 promotes proliferation and cisplatin resistance of oral squamous cell carcinoma by suppressing miR-184 expression. Cancer Med. 2017;6(12):2897–2908. doi:10.1002/cam4.2017.6.issue-12
28. Zhou Y, Wang X, Zhang J, et al. Artesunate suppresses the viability and mobility of prostate cancer cells through UCA1, the sponge of miR-184. Oncotarget. 2017;8(11):18260–18270. doi:10.18632/oncotarget.15353
29. Emdad L, Janjic A, Alzubi MA, et al. Suppression of miR-184 in malignant gliomas upregulates SND1 and promotes tumor aggressiveness. Neuro-Oncology. 2015;17(3):419–429. doi:10.1093/neuonc/nou220
30. Yuan Q, Gao W, Liu B, Ye W. Upregulation of miR-184 enhances the malignant biological behavior of human glioma cell line A172 by targeting FIH-1. Cell Physiol Biochem. 2014;34(4):1125–1136. doi:10.1159/000366326
31. Gao B, Gao K, Li L, Huang Z, Lin L. miR-184 functions as an oncogenic regulator in hepatocellular carcinoma (HCC). Biomed Pharmacother. 2014;68(2):143–148. doi:10.1016/j.biopha.2013.09.005
32. Wong TS, Liu XB, Wong BY, et al. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14(9):2588–2592. doi:10.1158/1078-0432.CCR-07-0666
33. Li H, Chen S, Liu J, et al. Long non-coding RNA PVT1-5 promotes cell proliferation by regulating miR-126/SLC7A5 axis in lung cancer. Biochem Biophys Res Commun. 2018;495(3):2350–2355. doi:10.1016/j.bbrc.2017.12.114
34. Wu J, Zhao J, Sun L, et al. Long non-coding RNA H19 mediates mechanical tension-induced osteogenesis of bone marrow mesenchymal stem cells via FAK by sponging miR-138. Bone. 2018;108:62–70. doi:10.1016/j.bone.2017.12.013
35. Chen H, Wang X, Yan X, et al. LncRNA MALAT1 regulates sepsis-induced cardiac inflammation and dysfunction via interaction with miR-125b and p38 MAPK/NFkappaB. Int Immunopharmacol. 2018;55:69–76. doi:10.1016/j.intimp.2017.11.038
36. Ando H, Natsume A, Iwami K, et al. A hypoxia-inducible factor (HIF)-3alpha splicing variant, HIF-3alpha4 impairs angiogenesis in hypervascular malignant meningiomas with epigenetically silenced HIF-3alpha4. Biochem Biophys Res Commun. 2013;433(1):139–144. doi:10.1016/j.bbrc.2013.02.044
37. Choudhry H, Harris AL, McIntyre A. The tumour hypoxia induced non-coding transcriptome. Mol Aspects Med. 2016;47-48:35–53. doi:10.1016/j.mam.2016.01.003
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.