Back to Journals » Cancer Management and Research » Volume 12
GSTM3 Function and Polymorphism in Cancer: Emerging but Promising
Authors Wang S , Yang J, You L, Dai M, Zhao Y
Received 16 July 2020
Accepted for publication 23 September 2020
Published 20 October 2020 Volume 2020:12 Pages 10377—10388
DOI https://doi.org/10.2147/CMAR.S272467
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Shunda Wang, Jinshou Yang, Lei You, Menghua Dai,* Yupei Zhao*
Department of General Surgery, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Menghua Dai, Yupei Zhao
Department of General Surgery, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100730, People’s Republic of China
Tel/Fax +86-10-6915-2600
Email [email protected]; [email protected]
Abstract: Cancer is a major cause of human mortality; however, the molecular mechanisms and proteomic biomarkers that cause tumor progression in malignant tumors are either unknown or only partially revealed. Glutathione S-transferases mu3 (GSTM3), which belongs to a family of xenobiotic detoxifying phase II enzymes, is associated with carcinogen detoxification and the metabolism of exogenous electrophilic substances. It has been reported that GSTM3 has different polymorphisms in various tumor cells and regulates tumorigenesis, cell invasion, metastasis, chemoresistance, and oxidative stress. Deep research into the regulatory mechanisms involved in disorders of GSTM3 expression and the function of GSTM3 in different cancers may facilitate improvements in cancer prevention and targeted therapy. The combination of GSTM3 with other family members can regulate the carcinogenesis and susceptibility to different cancers in humans. GSTM3 also regulates the reactive oxygen species (ROS) and participates in oxidative stress-mediated pathology. Here, we provide a general introduction to GSTM3 in order to better understand the role of GSTM3 in cancer.
Keywords: GSTM3, cancer, polymorphism, ROS
Introduction
Cancer remains a critical cause of human mortality. It is reported that globally there will be approximately 22 million new malignant tumor diagnoses and 13 million mortalities each year by the 2030s.1 Carcinogenesis maybe caused by variants in genetic levels, and in turn, genetic factors can influence the incidence rate. Glutathione S-transferases (GSTs) include a family of xenobiotic detoxifying phase II enzymes which mainly catalyze the connection of glutathione to a variety of electrophilic chemical compounds, which are involved in carcinogen detoxification and the metabolism of various bioactive compounds.2 GST families are highly polymorphic, and allele mutations or genetic deletions of a certain base lead to a predisposition for development of human cancer.3 Eight different classes of these cytoplasmic mammalian GSTs have already been identified: mu, pi, alpha, omega, kappa, sigma, zeta, and theta. The most widely studied families are mu (GSTM), pi (GSTP), alpha (GSTA), and theta (GSTT).4
The genes which encode GST mu3 are located on chromosome 1p13.3 and are known to be highly polymorphic.5 At present, GST mu3 is known to have two alleles: GSTM3*A and GSTM3*B. Like the GST family, the GSTM3 gene exerts an important function in the detoxification of the chemical substrates or electronic compounds, such as toxins, carcinogens, and production of oxidative stress.6,7 The relationship between the GST family and cancer has been widely explored, but the exact role of the GSTM3 gene has not been fully elucidated. The GSTM3 gene has been shown to significantly regulate individual susceptibility to cancers, such as laryngeal cancer, urinary cancer, and breast cancer.8–11 Contradictory conclusions have been published previously regarding the connection between polymorphic GSTM3 and protective or risky results in different malignant neoplasms. This is the first review of GSTM3 and it aims to provide a comprehensive overview of the functional expression and polymorphism of the GSTM3 gene in oral and esophageal carcinomas, prostate carcinoma, renal cancer, lung cancer, and other cancers. Additionally, it aims to briefly describe the molecular mechanisms involved in the progression of malignant disease.
GSTM3 Gene Function
It has been confirmed that the GSTM3 gene exerts an important role in the metabolism of toxic agents, such as the polyaromatic hydrocarbon, benzo (a) pyrene, that is released from tobacco. GSTM3 has overlapping substrate specificity with GSTM1.2 Each GST gene from each class is totally clustered on the identical chromosome.4 The GSTM1–GSTM5 genes are located on chromosome 1p and occupy a length of approximately 100 kb. The genes which encode the mu class 3 of GST are reported to be localized on 1p chromosome 13.3. The most common subtype is GSTM3*A. On the other hand, GSTM3*B has a 3-base pair deletion in intron 6, which produces a recognition binding site for yin yang 1(YY1), regarded as the transcription factor assumed to regulate the GSTM3 expression (Figure.1). YY1, a zinc finger transcription factor, can either activate or repress transcription with different efficiencies in the metabolism of carcinogens.12 GSTM3 also shares an amino acid sequence identity of about 70% with members of the mu class and less than 35% sequence identity with the alpha, pi, and theta classes, indicating that the GSTM3 enzyme functions in the detoxification of carcinogenic compounds.13 Moreover, it shows a linkage disequilibrium with the GSTM1 genotype.5 It can change the three-dimensional structure and make this region more susceptible to DNA-conjugated proteins, thus affecting the genetic regulation of other GSTM genes. Studies have indicated that changes in the expression level of GSTM3 have been observed in renal cancer, colon cancer, breast cancer, cervical cancer, and other cancers.14–18 Moreover, except for various neoplasms, GSTM3 is also reported to exert an important function in other kinds of diseases, such as Alzheimer’s disease, cataracts, neurocysticercosis, cystic fibrosis, and Brugada syndrome.19–24
 |
Figure 1 Gene cluster of GSTM1-5 arranged as 5ʹ-GSTM4-M2-M1-M5-M3-3ʹ and mutations of GSTM3 on chromosome 1p13. 3. |
In addition, GSTM3 presents an ethnic polymorphism frequency. The homozygous wild type (GSTM3*A) is more common than the homozygous type (GSTM3*B).25 In a Caucasian population from the United Kingdom, the frequency of GSTM3 polymorphism was 71% for the homozygous wild type (GSTM3*A) and only 4% for the homozygous type (GSTM3*B).5 Furthermore, such frequencies were also shown in people from Italy, Romania, and Sweden.25 However, in Asian countries, the polymorphic rate of wild type (GSTM3*A) was up to nearly 8289% in India and Malaysia.26,27 Moreover, the GSTM3*B genotype was more frequently observed in Africans compared to Europeans.28,29 Besides, the polymorphism of GSTM3 has also been reported to act as a hazard element for malignant tumors, but with conflicting consequences in different races, which might be aroused by a homozygous deletion of the GSTM3 gene.30,31
The Role of GSTM3 in Cancers
Cancers in the Urinary System
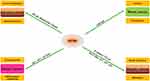
GSTM3 has been reported as being dysregulated in various cancers and the protein expression of GSTM3 has been reported to exert a peculiar function in the progression or inhibition of tumors (Figure 2). In renal clear cell carcinoma (RCC), Wang32 showed that GSTM3 rs1055259 was significantly associated with susceptibility to RCC. It was also verified to modify the GSTM3 protein synthesis by blocking miR-556 binding, then reducing the activity of reactive oxygen species (ROS) and the progression of RCC. Furthermore, Tan15 found that the upregulation of GSTM3 decreased the ability of anchorage-independent growth in renal cancer cells, which clearly proved that GSTM3 serves as a tumor suppressor in RCC. Furthermore, a case–control study showed that the C allele of GSTM3-rs1332018 predicted poor prognosis. The immunohistochemistry revealed different GSTM3 expression patterns in nephrons. In addition, global analysis of gene expression in RCC specimens also showed that GSTM3 varied in different parts of the tissue.33
Cancers in the Reproductive System
As for prostate cancer (PC), a bioinformatics analysis using the Gene Expression Omnibus database showed that the dysregulation between hsa-let-7a-5p/CDKN1A and hsa-miR-92b-3p/GSTM3 pairs was associated with platinum-based chemoresistance of metastatic PC.34 The GSTM3 rs7483 polymorphism may be a promising biomarker for prostate cancer patients treated with androgen-deprivation therapy.35 In metastatic PC, GSTM3 rs7483 was associated with significant risk of progression. In non-metastatic PC, the AG/GG allele in GSTM3 rs7483 was associated with a lower risk of progression to castration-resistant prostate cancer. In terms of cervical cancer, Alberto16 pointed out that GSTM3 influenced cell maintenance, cell apoptosis, and the intracellular stress response through the NF-κB and MAPK/ERK signal pathway. In HeLa cells, GSTM3 interaction with TRAF6 was predicted by the SysBiomics databases,36 then identified by co-immunoprecipitation. Recently, Lollo also pointed out that GSTM3 acted as a key epithelial–mesenchymal transition inducer operating in amniotic epithelial cells using RNA sequencing.37
Cancers in the Digestive System
As for hepatoma, GSTM3 plays a vital role in reversing the radio-resistance of hepatic cell carcinoma (HCC). It could also be an effective therapeutic target because it makes HCC cells sensitive to radiotherapy. GSTM3 also regulates the expression of Bcl-2, Bax, p21, p27, and p53, which are related to the cell cycle and cell apoptosis.38 The level of GSTM3 mRNA expression in HCC cells showed a significant connection with the process of cellular detoxification, which also prompts the metastatic ability in cancer cells. GSTM3 might be a new downstream gene of the β-catenin/Tcf-Lef pathway in hepatic cancer cells.39 Hepatitis is also known to correlate with hepatic cancer. Qi40 reported that the methylation of GSTM3 promoter was associated with oxidative stress-related liver failure. Furthermore, it could also influence the severity of liver damage in patients with acute-on-chronic hepatitis B. Then, Sun41 found that the low methylation level of GSTM3 promoter predicted good prognosis of hepatitis B liver failure. In colon cancer, Stephan14 used tissue-based proteomics to show a strong association between gene expression level and metastatic lymph nodes. The over-expression of GSTM3 reduced the overall survival rate of patients.18 As for chemotherapy, metastatic colon cancer cells treated with cisplatin demonstrated an increasing level of GSTM3 expression than cells without cisplatin treatment.42
Other Cancers
Currently, the exact function of GSTM3 in breast cancer is still unclear. It has been reported that the mRNA expression level is high in HER2-positive or ER-positive cancer types. Estrogen could stimulate ER to recruit the SP1, AP-1, and EP 300 transcription factors to bind to the GSTM3 promoter, resulting in upregulation of the GSTM3 gene in ER-positive breast cancer.43,44 On the other hand, ERα-mediated GSTM3 downregulation could prevent the H2O2-induced cytotoxicity and enhance the carcinogenic process of breast cancer. Long-term tamoxifen treatment for breast carcinoma has been reported to reduce levels of GSTM3 and enhance the sensitivity to H2O2.44 Besides, Asik found that GSTM3, together with another phase II enzyme GSTZ1, were increased under the high concentrations of 2-amino-2-deoxy-glucose treatment in breast cancer cells.45 Besides, when it comes to glioma, Li46 showed that GSTM3 had a structure to integrate with long LncRNA growth-arrest specific transcript 5 and downregulated GSTM3, inhibiting the proliferation, invasion, and migration, but exerting apoptosis and enhancing oxidative stress.
GSTM3 Gene Polymorphism in Cancer
Single-nucleotide polymorphisms (SNPs) of the cancer-encoding gene have been identified to alter the genetic expression and influence the morbidity of cancers in different individuals. Genotyping of the important alleles for the gene has been associated with cancer risk, and there is a strong linkage between a kind of polymorphism and increasing progression of certain cancers. Several studies have indicated that the SNPs of GSTM3 could change the activity of enzymes, which modify the effect on personal sensitivity to toxins or pathologies.47 Indeed, there are plenty of studies that have affirmed that the polymorphisms of GSTM3 are strongly associated with carcinogenesis and tumor progression15,29,32 (Table 1).
 |
Table 1 General List of the GSTM3 Polymorphism and Cancer Risk in Different Types of Malignant Tumors |
HNC Cancer
Head and neck cancer (HNC)—including cancers of the oral cavity, pharynx, and larynx—is the sixth most common malignant tumor globally.48 Cha49 demonstrated that the overexpression of the GSTM3*B allele seemed to be associated with an increased risk of laryngeal squamous carcinoma. Nadejda50 collected peripheral blood samples from 129 patients and 172 healthy controls. Then, he verified the linkage between the cancer risk and the polymorphisms of GSTM3. The results showed that patients with the GSTM3 (AB or BB) genotype had a two-fold risk of larynx cancer than healthy individuals. However, Matthias51 found a reduced frequency of GSTM3 (AB or BB) alleles in larynx cancer patients compared to healthy people. As for esophageal cancer, Jain8 reported that patients with the heterozygous genotype GSTM3*AB had an increased risk of esophageal cancer. Moreover, GSTM3 polymorphism might modulate the risk of adenocarcinoma and the region of the tumors. Besides, Nilabja52 reported that smokers with the GSTM3*A genotype were at higher risk of oral cancer than controls. Furthermore, in 2005, Mousumi substantiated the study by Nilabja with expanded sample sizes. The genotype GSTM3*A increased the susceptibility of oral cancer among smokers.53 However, Rao failed to observe a relationship between GSTM3 polymorphism and risk of developing oral carcinoma.54 A meta-analysis by Xu drew the conclusion that the GSTM3*A/B could act as a crucial protective genotype for HNC, especially for laryngeal cancer.55
Lung Cancer
GSTM3 has been reported to express inconsistently in lung tissue, and this gene has the ability to increase the susceptibility to lung cancer.56–58 Sisko56 showed that overexpression of this gene might lead to development of adenocarcinoma in the lungs of GSTM1 null individuals. However, GSTM3 was not influenced by the GSTM1 genotype in long-term smokers. In addition, Figueras59 noticed that the combined genotype between GSTM1 null and GSTM3*AA might increase the lung cancer risk. Moreover, Risch57 showed that the risky allele for GSTM3 conferring reduced enzyme activity was present at high levels in squamous cell carcinomas. This result was in accordance with a decreased detoxication of carcinogenic substances from cigarette smoke that tends to lead to development of squamous cell carcinomas. On the contrary, the meta-analysis performed by Feng in 2012 showed little connection between GSTM3 polymorphism and the risk of lung cancer.60 Sissung showed that GSTM3 rs7483 was associated with paclitaxel progression-free survival in lung cancer patients.61 Therefore, the correlation between GSTM3 polymorphism and the risk of lung cancer still needs to be explored.
Prostate Cancer
The mutation of the GSTM3 gene has also been shown to enhance the risk of cancer formation in the urogenital system, particularly the susceptibility to prostate carcinogenesis. Pravin analyzed the GSTM3 intron 6 polymorphisms among Indian patients with PC and pointed out that the GSTM3 (AB + BB) genotype took part in the evolvement of PC.62 And the risk was increased in cigarette smokers and alcohol consumers. Another study from Medeiros showed the function of different alleles of GSTM3 in PC patients in Europe, and the polymorphism in the GSTM3 might be an important biomarker for PC risk.63 Moreover, GSTT1 null was reported to be overrepresented in advanced PC patients. The carriers of the GSTM3*BB genotype are reported to have a five-fold increased risk of PC. GSTM3*B, regarded as an at-risk allele, increases the transcriptional potentiality, and strengthens the detoxification ability of GSTM3-encoding protein.64 The above evidence could demonstrate that carriers of GSTM3 variant genotype apparently played an important function in mediating susceptibility for PC. Larger-scale molecular investigations are needed to verify the role of GSTM3 genetic polymorphisms in PC.
Other Cancers
GSTM3 has also been investigated in other malignant tumors because of its polymorphism. As for bladder cancer, Schnakenberg9 proved that the intron 6 mutation of GSTM3 enhanced the susceptibility to bladder carcinoma. And Hariom65 showed that GSTM3*AB genotypes might increase the risk of cervical cancers. Moreover, Yu66 pointed out that SNP of the GSTM3 gene cluster could contribute to the carcinogenesis of breast cancer. And the catalyzing ability of GSTM3 in normal tissue could act as a protective factor against the risk of breast carcinoma. However, Jaramillo-Rangel67 proved that GSTM3 has no association with breast cancer susceptibility. In colorectal carcinoma, the interactions of polymorphic genotypes of GSTM3 influenced individual risk and the absence of the GSTM3*B variant was a protective factor, especially its association with the GSTM1 genotype.68 However, another case–control study by Cortessis reported that GSTM3 had no correlation with the risk of colorectal adenomas.69 One meta-analysis concluded that the polymorphisms in GSTM3 (AA versus BB) and osteosarcoma risk were significantly correlated.70 An observational study in North America demonstrated that GSTM3 also contributed to increased susceptibility to meningioma and glioma,71 but the same conclusion was not verified in residents of Europe.72 Besides, Yengi also showed that GSTM3 polymorphism interactions with cytochrome P450 served as risk factors for multiple cutaneous basal cell carcinoma.64 Taken together, no one can ignore the association between GSTM3 polymorphism and tumorigenesis. The exact role of GSTM3 polymorphism and its underlying mechanisms in the suppressibility of cancer still need to be explored in larger studies.
GSTM3 Interaction with Other GSTs in Carcinogenesis
The correlation of different genotypes among the GST gene family influences the individual risk of tumorigenesis. Besides GSTM3, many researchers have paid attention to the interaction between genetic variants in other GSTM family genes. The co-function of GSTM1 and GSTM3 has been widely investigated. Genetic variants in the GSTM3 gene depend on GSTM1. Some studies have also pointed out that the GSTM3 genotype might regulate the effects of GSTM1.11 In breast cancer, the incidence of breast cancer risk was probably caused by haplotypes of the GSTM3 gene cluster in the absence of GSTM1.66 Another study proved the susceptibility to breast cancer among premenopausal women could increase if they simultaneously expressed the GSTM3*B and the GSTP1 genotypes. On the other hand, it also mentioned that the risk would increase fiercely if they lacked the GSTT1 at the same time.10 In colon cancer, the absence of GSTM3*B was a protective factor, especially in accordance with the null type of GSTM1. The genotype analysis of patients with distal and proximal tumors revealed that the constitution of the GSTM3*B allele and GSTM1-null were the most frequent genotypes.68 In bladder cancer, individuals without the homozygous type of GSTM1 or GSTM3 were apparently sensitive to carcinogenesis.9 Besides, the “wild types” of GSTM3 or GSTM1 served as protective genotypes for bladder cancer. Taken together, this infers that GSTM3 can influence the incidence and risk of cancer solely or in combination with other genes such as GSTT or the GSTP family. However, because some personal data were absent in this review, we could not conduct more detailed analyses. We also analyzed the inner linkage of effects of GSTM3 with other risk factors. Even though the present conclusions for gene-gene analysis are not homogeneous, they obviously advocate that GSTM3 functions as a risk regulator, either individually or in cooperation with other polymorphic genes.
GSTM3 as a Prognostic Indicator in Pan-Cancer
The overall survival curves of GSTM3 are depicted in an online Kaplan–Meier (KM) plotter (www.kmplot.com).73 This is a novel interactive website that evaluates gene expression and overall survival (OS) according to TCGA. Each type of cancer corresponds to a different group of people. Median values for each cancer were selected as cutoff values to draw the KM plot. The connection between the GSTM3 mRNA level and the OS of cancer patients is shown in (Figure 3). From the available data, GSTM3 was shown to correlate significantly with OS in six malignancies (405 cases of bladder cancer, 371 cases of live hepatocellular carcinoma, 165 cases of rectal adenocarcinoma, 288 cases of renal clear cell carcinoma, 304 cases of cervical squamous cancer, 1090 cases of breast cancer). On the one hand, the high level of GSTM3 was significantly associated with poor OS in bladder cancer (P=0.033), live hepatocellular carcinoma (P=0.0033), and rectal adenocarcinoma (P=0.0029), indicating its tumor-promoting role. On the other hand, it serves as a tumor suppressor in RCC (P=0.00049), cervical squamous cancer (P=0.041), and breast cancer (0.027). The above results demonstrated that GSTM3 plays versatile roles in malignant tumors. Further exploration is much anticipated in terms of analyses of the differentiation, gender, age, and pathological stage on the prognostic value of GSTM3. Additionally, these results are derived from public databases and need to be verified by further studies. Due to the small sample size, the conclusions here should be regarded as preliminary or tentative until they are verified in extensive population-based research.
GSTM3 in ROS and Oxidative Stress
Oxidative stress reflects an imbalance between the production of ROS and antioxidant defense systems.74 It exerts an important role in the pathological process of malignant tumors, hypertension, diabetes, and even neurodegenerative diseases.75–77 It is a conflict between antioxidant and oxidant balance, which results in cellular injury, and the imbalance may be caused by excessive ROS.78,79 ROS include superoxide anion, hydrogen peroxide, hydroxyl radical, amongst others, and are produced naturally by the activity of the mitochondrial electron transport chain.80 It is known that cells have defense mechanisms to break down ROS by synthesis proteins binding to ROS and composing hydrophilic chemicals that are readily secreted. The antioxidant defense system can protect DNA from the damaging effects of oxidative stress.81 The GST family appears to be very significant in preventing cells from harmful endogenous substances.82 Particularly, GSTM3 could protect cells against electrophilic damage via catalyzing the conjugation of ROS and glutathione, playing a pivotal role in detoxification and ROS clearance.83,84
In a former study, Wang32 reported that GSTM3 served as a tumor suppressor by modulating the activity of ROS. The variant G allele of rs1055259 might elevate GSTM3 expression by affecting the binding of miR-556 and 3ʹUTR of the GSTM3 gene, resulting in low ROS levels and high thiol levels, and further suppressing the proliferation and invasion of RCC cells. Antioxidant capacity has been proven to be associated with GSTM3 regulation in RCC. Augmented ROS can initiate redox-linked signaling responses and irreversible injuries in RCC.85,86 Alteration of cellular redox balance in RCC might also be correlated with the expression of GST enzymes.85 Shiota found that GSTM3 overexpression was accompanied by elevated ROS accumulation in castration-resistant PC cells. The polymorphism in GSTM3 alters the activity of antioxidant enzymes to contribute to hormonal resistance therapy through oxidative stress.35 Besides, Li46 found that downrated GSTM3 enhanced the production of ROS in glioma cells. Recently, GSTM3 was proven to play a crucial role in regulation of the oxidative stress and mitochondrial function in mammalian sperm.87 Polymorphisms in antioxidant genes and the gene–gene interactions, such as GSTM3 rs3814309/NQO1, may increase the potential risk of male infertility in the Chinese population.88 Mechanically, Chu89 pointed out that the domain-containing protein 1 (NSD1) regulates the expression of GSTM3 in response to oxidative stress. NSD1 was associated with oxidative stress by H2O2-induced. NSD1 suppression led to the reduction of GSTM3 levels through the −63A/C TATA box. Interestingly, Liu90 provided strong proof that GSTM3−63A/C served as a potential target responding to oxidative stress. In a word, glutathione S-transferase mu3 has a strong antioxidant ability by resisting numerous harmful substances in the environment.
Conclusions and Prospects
As a significant component of the mu-class subfamily, GSTM3 exerts an important role in protecting cancer cells from oxidative stress and helps to prevent against carcinogens. It is also closely associated with cancer susceptibility and tumorigenesis as a result of polymorphism or aberrant expression. At the proteomic level, GSTM3 is closely involved in cell proliferation, tumor invasion, metastasis, chemoresistance, and even prognosis in different kinds of cancers. It also functions as an important intracellular radical scavenger and protects cells against ROS. At the genetic level, genotyping of important alleles for GSTM3 gene mutations may lead to increased cancer risk. Polymorphisms exert an impact on individual sensitivity to certain pathologies.
In conclusion, the present article is the first comprehensive review of GSTM3 and discusses the mechanisms of GSTM3 in different malignancies. We systematically reviewed the biological functions and genetic polymorphisms of GSTM3 among pan-cancer and showed the promising future application of manipulating GSTM3 expression to improve clinical outcomes in patients with malignancy. Furthermore, the identification of the underlying mechanism can evolve into the foundations of therapeutic targets. In the field of research into GSTM3 and malignancies, convictive animal experiments and clinical studies are still insufficient to illustrate its role in the treatment of malignant diseases. Finally, the molecular mechanisms of GSTM3 in different cancers warrant further study. We strongly believe that GSTM3 could be a potential target in the future.
Acknowledgments
We thank all the staff from Department of General Surgery, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The project was funded by Chinese Academy of Medical Science (CAMS) Innovation Fund for Medical Sciences (Grant No. 2016-I2M-3-005). Another supportive source of this work was from the National Major Research and Development Programs of the Ministry of Science and Technology of the People’s Republic of China during the 13th Five-Year Plan Period (Grant No. 2017YFC1308602).
Disclosure
All of the authors declare no conflicts of interest in this work.
References
1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359386. doi:10.1002/ijc.29210
2. Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30(6):445–600.
3. Brooks JD, Weinstein M, Lin X, et al. CG island methylation changes near the GSTP1 gene in prostatic intraepithelial neoplasia. Cancer Epidemiol Biomarkers Prev. 1998;7(6):531–536.
4. Hayes JD, Strange RC. Glutathione S-transferase polymorphisms and their biological consequences. Pharmacology. 2000;61(3):154–166. doi:10.1159/000028396
5. Inskip A, Elexperu-Camiruaga J, Buxton N, et al. Identification of polymorphism at the glutathione S-transferase, GSTM3 locus: evidence for linkage with GSTM1*A. Biochem J. 1995;312(Pt 3):713–716. doi:10.1042/bj3120713
6. Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi:10.1146/annurev.pharmtox.45.120403.095857
7. Li D, Gao Q, Xu L, et al. Characterization of glutathione S-transferases in the detoxification of metolachlor in two maize cultivars of differing herbicide tolerance. Pestic Biochem Physiol. 2017;143:265–271. doi:10.1016/j.pestbp.2016.12.003
8. Jain M, Kumar S, Lal P, Tiwari A, Ghoshal UC, Mittal B. Role of GSTM3 polymorphism in the risk of developing esophageal cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(1):178–181. doi:10.1158/1055-9965.EPI-06-0542
9. Schnakenberg E, Breuer R, Werdin R, Dreikorn K, Schloot W. Susceptibility genes: GSTM1 and GSTM3 as genetic risk factors in bladder cancer. Cytogenet Cell Genet. 2000;91(14):234–238. doi:10.1159/000056851
10. Mitrunen K, Jourenkova N, Kataja V, et al. Glutathione S-transferase M1, M3, P1, and T1 genetic polymorphisms and susceptibility to breast cancer. Cancer Epidemiol Biomarkers Prev. 2001;10(3):229–236.
11. Pandey SN, Jain M, Nigam P, Choudhuri G, Mittal B. Genetic polymorphisms in GSTM1, GSTT1, GSTP1, GSTM3 and the susceptibility to gallbladder cancer in North India. Biomarkers. 2006;11(3):250–261. doi:10.1080/13547500600648697
12. Flanagan JR. Autologous stimulation of YY1 transcription factor expression: role of an insulin-like growth factor. Cell Growth Differ. 1995;6(2):185–190.
13. Patskovsky YV, Huang MQ, Takayama T, Listowsky I, Pearson WR. Distinctive structure of the human GSTM3 gene-inverted orientation relative to the mu class glutathione transferase gene cluster. Arch Biochem Biophys. 1999;361(1):85–93. doi:10.1006/abbi.1998.0964
14. Meding S, Balluff B, Elsner M, et al. Tissue-based proteomics reveals FXYD3, S100A11 and GSTM3 as novel markers for regional lymph node metastasis in colon cancer. J Pathol. 2012;228(4):459–470. doi:10.1002/path.4021
15. Tan X, Wang Y, Han Y, et al. Genetic variation in the GSTM3 promoter confer risk and prognosis of renal cell carcinoma by reducing gene expression. Br J Cancer. 2013;109(12):3105–3115. doi:10.1038/bjc.2013.669
16. Checa-Rojas A, Delgadillo-Silva LF, Velasco-Herrera MDC, et al. GSTM3 and GSTP1: novel players driving tumor progression in cervical cancer. Oncotarget. 2018;9(31):21696–21714. doi:10.18632/oncotarget.24796
17. Ye Z, Song H, Higgins JP, Pharoah P, Danesh J. Five glutathione s-transferase gene variants in 23,452 cases of lung cancer and 30,397 controls: meta-analysis of 130 studies. PLoS Med. 2006;3(4):e91. doi:10.1371/journal.pmed.0030091
18. Mitra AP, Pagliarulo V, Yang D, et al. Generation of a concise gene panel for outcome prediction in urinary bladder cancer. J Clin Oncol. 2009;27(24):3929–3937. doi:10.1200/JCO.2008.18.5744
19. Bullock JM, Medway C, Cortina-Borja M, et al. Discovery by the epistasis project of an epistatic interaction between the GSTM3 gene and the HHEX/IDE/KIF11 locus in the risk of Alzheimer’s disease. Neurobiol Aging. 2013;34(4):1309e13011307. doi:10.1016/j.neurobiolaging.2012.08.010
20. Li B, Zhou J, Zhang G, et al. Relationship between the altered expression and epigenetics of GSTM3 and age-related cataract. Invest Ophthalmol Vis Sci. 2016;57(11):4721–4732. doi:10.1167/iovs.16-19242
21. Maes OC, Schipper HM, Chong G, Chertkow HM, Wang E. A GSTM3 polymorphism associated with an etiopathogenetic mechanism in Alzheimer disease. Neurobiol Aging. 2010;31(1):34–45. doi:10.1016/j.neurobiolaging.2008.03.007
22. Singh A, Prasad KN, Singh AK, et al. Human glutathione s-transferase enzyme gene polymorphisms and their association with neurocysticercosis. Mol Neurobiol. 2017;54(4):2843–2851. doi:10.1007/s12035-016-9779-4
23. Magalhaes M, Rivals I, Claustres M, et al. DNA methylation at modifier genes of lung disease severity is altered in cystic fibrosis. Clin Epigenetics. 2017;9:19. doi:10.1186/s13148-016-0300-8
24. Juang JJ, Binda A, Lee SJ, et al. GSTM3 variant is a novel genetic modifier in Brugada syndrome, a disease with risk of sudden cardiac death. E Bio Medicine. 2020;57:102843. doi:10.1016/j.ebiom.2020.102843
25. Mozzoni P, De Palma G, Scotti E, Capelletti M, Mutti A. Characterization of GSTM3 polymorphism by real-time polymerase chain reaction with LightCycler. Anal Biochem. 2004;330(1):175–177. doi:10.1016/j.ab.2004.03.006
26. Alshagga MA, Mohamed N, Nazrun Suhid A. Frequencies of glutathione s-transferase (GSTM1, GSTM3 AND GSTT1) polymorphisms in a Malaysian population. Arch Med Sci. 2011;7(4):572–578. doi:10.5114/aoms.2011.24123
27. Buch SC, Notani PN, Bhisey RA. Polymorphism at GSTM1, GSTM3 and GSTT1 gene loci and susceptibility to oral cancer in an Indian population. Carcinogenesis. 2002;23(5):803–807. doi:10.1093/carcin/23.5.803
28. Garte S, Gaspari L, Alexandrie AK, et al. Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol Biomarkers Prev. 2001;10(12):1239–1248.
29. Tetlow N, Robinson A, Mantle T, Board P. Polymorphism of human mu class glutathione transferases. Pharmacogenetics. 2004;14(6):359–368. doi:10.1097/00008571-200406000-00005
30. Rebbeck TR. Molecular epidemiology of the human glutathione S-transferase genotypes GSTM1 and GSTT1 in cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 1997;6(9):733–743.
31. Strange RC, Fryer AA. The glutathione S-transferases: influence of polymorphism on cancer susceptibility. IARC Sci Publ. 1999;(148):231–249.
32. Wang Y, Yang ZY, Chen YH, et al. A novel functional polymorphism of GSTM3 reduces clear cell renal cell carcinoma risk through enhancing its expression by interfering miR-556 binding. J Cell Mol Med. 2018;22(6):3005–3015. doi:10.1111/jcmm.13528
33. Tan X, Zhai Y, Chang W, et al. Global analysis of metastasis-associated gene expression in primary cultures from clinical specimens of clear-cell renal-cell carcinoma. Int J Cancer. 2008;123(5):1080–1088. doi:10.1002/ijc.23637
34. Ma H, Wang LY, Yang RH, Zhou Y, Zhou P, Kong L. Identification of reciprocal microRNA-mRNA pairs associated with metastatic potential disparities in human prostate cancer cells and signaling pathway analysis. J Cell Biochem. 2019;120(10):17779–17790. doi:10.1002/jcb.29045
35. Shiota M, Fujimoto N, Itsumi M, et al. Gene polymorphisms in antioxidant enzymes correlate with the efficacy of androgen-deprivation therapy for prostate cancer with implications of oxidative stress. Ann Oncol. 2017;28(3):569–575. doi:10.1093/annonc/mdw646
36. Rouillard AD, Gundersen GW, Fernandez NF, et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database. 2016;2016.
37. Di Lollo V, Canciello A, Orsini M, et al. Transcriptomic and computational analysis identified LPA metabolism, KLHL14 and KCNE3 as novel regulators of Epithelial-Mesenchymal Transition. Sci Rep. 2020;10(1):4180. doi:10.1038/s41598-020-61017-y
38. Sun Y, Wang Y, Yin Y, Chen X, Sun Z. GSTM3 reverses the resistance of hepatoma cells to radiation by regulating the expression of cell cycle/apoptosis-related molecules. Oncol Lett. 2014;8(4):1435–1440. doi:10.3892/ol.2014.2358
39. Li YS, Liu M, Nakata Y, Tang HB. Beta-catenin accumulation in nuclei of hepatocellular carcinoma cells up-regulates glutathione-s-transferase M3 mRNA. World J Gastroenterol. 2011;17(13):1772–1778. doi:10.3748/wjg.v17.i13.1772
40. Qi L, Zou ZQ, Wang LY, et al. Methylation of the glutathione-S-transferase M3 gene promoter is associated with oxidative stress in acute-on-chronic hepatitis B liver failure. Tohoku J Exp Med. 2012;228(1):43–51. doi:10.1620/tjem.228.43
41. Sun FK, Gao S, Fan YC, et al. High promoter methylation levels of glutathione-S-transferase M3 predict poor prognosis of acute-on-chronic hepatitis B liver failure. Hepatol Res. 2017;47(6):566–573. doi:10.1111/hepr.12777
42. Huerta S, Harris DM, Jazirehi A, et al. Gene expression profile of metastatic colon cancer cells resistant to cisplatin-induced apoptosis. Int J Oncol. 2003;22(3):663–670.
43. Bieche I, Girault I, Urbain E, Tozlu S, Lidereau R. Relationship between intratumoral expression of genes coding for xenobiotic-metabolizing enzymes and benefit from adjuvant tamoxifen in estrogen receptor alpha-positive postmenopausal breast carcinoma. Breast Cancer Res. 2004;6(3):R252263. doi:10.1186/bcr784
44. Lin JH, Tu SH, Chen LC, et al. Oestrogen receptor-regulated glutathione S-transferase mu 3 expression attenuates hydrogen peroxide-induced cytotoxicity, which confers tamoxifen resistance on breast cancer cells. Breast Cancer Res Treat. 2018;172(1):45–59. doi:10.1007/s10549-018-4897-5
45. Asik E, Aslan TN, Volkan M, Guray NT. 2-Amino-2-deoxy-glucose conjugated cobalt ferrite magnetic nanoparticle (2DG-MNP) as a targeting agent for breast cancer cells. Environ Toxicol Pharmacol. 2016;41:272–278. doi:10.1016/j.etap.2015.12.004
46. Li G, Cai Y, Wang C, Huang M, Chen J. LncRNA GAS5 regulates the proliferation, migration, invasion and apoptosis of brain glioma cells through targeting GSTM3 expression. The effect of LncRNA GAS5 on glioma cells. J Neurooncol. 2019;143(3):525–536. doi:10.1007/s11060-019-03185-0
47. Ouerhani S, Ben Bahria I, Rouissi K, Cherni L. Distribution of xenobiotic metabolising enzyme genotypes in different Tunisian populations. Ann Hum Biol. 2017;44(4):366–372. doi:10.1080/03014460.2016.1272714
48. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi:10.3322/caac.20107
49. Chatzimichalis M, Xenellis J, Tzagaroulakis A, et al. GSTT1, GSTM1, GSTM3 and NAT2 polymorphisms in laryngeal squamous cell carcinoma in a Greek population. J Laryngol Otol. 2010;124(3):318–323. doi:10.1017/S002221510999154X
50. Jourenkova-Mironova N, Voho A, Bouchardy C, et al. Glutathione S-transferase GSTM3 and GSTP1 genotypes and larynx cancer risk. Cancer Epidemiol Biomarkers Prev. 1999;8(2):185–188.
51. Matthias C, Bockmuhl U, Jahnke V, et al. Polymorphism in cytochrome P450 CYP2D6, CYP1A1, CYP2E1 and glutathione S-transferase, GSTM1, GSTM3, GSTT1 and susceptibility to tobacco-related cancers: studies in upper aerodigestive tract cancers. Pharmacogenetics. 1998;8(2):91–100. doi:10.1097/00008571-199804000-00001
52. Sikdar N, Paul RR, Roy B. Glutathione S-transferase M3 (A/A) genotype as a risk factor for oral cancer and leukoplakia among Indian tobacco smokers. Int J Cancer. 2004;109(1):95–101. doi:10.1002/ijc.11610
53. Majumder M, Sikdar N, Paul RR, Roy B. Increased risk of oral leukoplakia and cancer among mixed tobacco users carrying XRCC1 variant haplotypes and cancer among smokers carrying two risk genotypes: one on each of two loci, GSTM3 and XRCC1 (Codon 280). Cancer Epidemiol Biomarkers Prev. 2005;14(9):2106–2112. doi:10.1158/1055-9965.EPI-05-0108
54. Rao AK, Parameswar P, Majumdar S, Uppala D, Kotina S, Vennamaneni NH. Evaluation of genetic polymorphisms in glutathione s-transferase theta1, glutathione s-transferase mu1, and glutathione s-transferase mu3 in oral leukoplakia and oral squamous cell carcinoma with deleterious habits using polymerase chain reaction. Int J Appl Basic Med Res. 2017;7(3):181–185. doi:10.4103/ijabmr.IJABMR_58_16
55. Xu Y, Wang J, Dong W. GSTM3 A/B polymorphism and risk for head and neck cancer: a meta-analysis. PLoS One. 2014;9(1):e83851. doi:10.1371/journal.pone.0083851
56. Anttila S, Luostarinen L, Hirvonen A, et al. Pulmonary expression of glutathione S-transferase M3 in lung cancer patients: association with GSTM1 polymorphism, smoking, and asbestos exposure. Cancer Res. 1995;55(15):3305–3309.
57. Risch A, Wikman H, Thiel S, et al. Glutathione-S-transferase M1, M3, T1 and P1 polymorphisms and susceptibility to non-small-cell lung cancer subtypes and hamartomas. Pharmacogenetics. 2001;11(9):757–764. doi:10.1097/00008571-200112000-00003
58. Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274(5286):430–432. doi:10.1126/science.274.5286.430
59. Corbella JT-FMGJG-CEPNBGMRGDJ. Polymorphism of glutathione S-transferase M3: interaction with glutathione S-transferase M1 and lung cancer susceptibility. Biomarkers. 2000;5(1):73–80. doi:10.1080/135475000230550
60. Feng X, Dong CQ, Shi JJ, Zhou HF, He W, Zheng BS. Lack of association of glutathione S-transferase M3 gene polymorphism with the susceptibility of lung cancer. Asian Pac J Cancer Prev. 2012;13(9):4465–4468.
61. Sissung TM, Rajan A, Blumenthal GM, et al. Reproducibility of pharmacogenetics findings for paclitaxel in a heterogeneous population of patients with lung cancer. PLoS One. 2019;14(2):e0212097. doi:10.1371/journal.pone.0212097
62. Kesarwani P, Singh R, Mittal RD. Association of GSTM3 intron 6 variant with cigarette smoking, tobacco chewing and alcohol as modifier factors for prostate cancer risk. Arch Toxicol. 2009;83(4):351–356. doi:10.1007/s00204-008-0343-5
63. Medeiros R, Vasconcelos A, Costa S, et al. Metabolic susceptibility genes and prostate cancer risk in a southern European population: the role of glutathione S-transferases GSTM1, GSTM3, and GSTT1 genetic polymorphisms. Prostate. 2004;58(4):414–420.
64. Yengi L, Inskip A, Gilford J, et al. Polymorphism at the glutathione S-transferase locus GSTM3: interactions with cytochrome P450 and glutathione S-transferase genotypes as risk factors for multiple cutaneous basal cell carcinoma. Cancer Res. 1996;56(9):1974–1977.
65. Singh H, Sachan R, Devi S, Pandey SN, Mittal B. Association of GSTM1, GSTT1, and GSTM3 gene polymorphisms and susceptibility to cervical cancer in a North Indian population. Am J Obstet Gynecol. 2008;198(3):303e301306. doi:10.1016/j.ajog.2007.09.046
66. Yu KD, Fan L, Di GH, et al. Genetic variants in GSTM3 gene within GSTM4-GSTM2-GSTM1-GSTM5-GSTM3 cluster influence breast cancer susceptibility depending on GSTM1. Breast Cancer Res Treat. 2010;121(2):485–496. doi:10.1007/s10549-009-0585-9
67. Jaramillo-Rangel G, Ortega-Martinez M, Cerda-Flores RM, Barrera-Saldana HA. Polymorphisms in GSTM1, GSTT1, GSTP1, and GSTM3 genes and breast cancer risk in northeastern Mexico. Genet Mol Res. 2015;14(2):6465–6471. doi:10.4238/2015.June.11.22
68. Loktionov A, Watson MA, Gunter M, Stebbings WS, Speakman CT, Bingham SA. Glutathione-S-transferase gene polymorphisms in colorectal cancer patients: interaction between GSTM1 and GSTM3 allele variants as a risk-modulating factor. Carcinogenesis. 2001;22(7):1053–1060. doi:10.1093/carcin/22.7.1053
69. Cortessis V, Siegmund K, Chen Q, et al. A case-control study of microsomal epoxide hydrolase, smoking, meat consumption, glutathione S-transferase M3, and risk of colorectal adenomas. Cancer Res. 2001;61(6):2381–2385.
70. Wang Z, Xu H, He M, Wu H, Zhu Y, Su Z. The association of glutathione S-transferase polymorphisms in patients with osteosarcoma: evidence from a meta-analysis. Eur J Cancer Care. 2015;24(3):417–424. doi:10.1111/ecc.12197
71. De Roos AJ, Rothman N, Brown M, et al. Variation in genes relevant to aromatic hydrocarbon metabolism and the risk of adult brain tumors. Neuro Oncol. 2006;8(2):145–155. doi:10.1215/15228517-2005-003
72. Schwartzbaum JA, Ahlbom A, Lonn S, et al. An international case-control study of interleukin-4Ralpha, interleukin-13, and cyclooxygenase-2 polymorphisms and glioblastoma risk. Cancer Epidemiol Biomarkers Prev. 2007;16(11):2448–2454. doi:10.1158/1055-9965.EPI-07-0480
73. Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. doi:10.1371/journal.pone.0082241
74. Scioli MG, Storti G, D’Amico F, et al. Oxidative stress and new pathogenetic mechanisms in endothelial dysfunction: potential diagnostic biomarkers and therapeutic targets. J Clin Med. 2020;9:6. doi:10.3390/jcm9061995
75. Mahalingaiah PK, Singh KP. Chronic oxidative stress increases growth and tumorigenic potential of MCF-7 breast cancer cells. PLoS One. 2014;9(1):e87371.
76. Jenner P. Oxidative stress in Parkinson’s disease. Ann Neurol. 2003;53(Suppl 3):
77. Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48(2):158–167. doi:10.1016/j.molcel.2012.09.025
78. Maiese K, Morhan SD, Chong ZZ. Oxidative stress biology and cell injury during type 1 and type 2 diabetes mellitus. Curr Neurovasc Res. 2007;4(1):63–71. doi:10.2174/156720207779940653
79. Kotsafti A, Scarpa M, Castagliuolo I, Scarpa M. Reactive oxygen species and antitumor immunity-from surveillance to evasion. Cancers. 2020;12:7. doi:10.3390/cancers12071748
80. Banerjee S, Ghosh S, Mandal A, Ghosh N, Sil PC. ROS-associated immune response and metabolism: a mechanistic approach with implication of various diseases. Arch Toxicol. 2020.
81. Moloney JN, Cotter TG. ROS signalling in the biology of cancer. Semin Cell Dev Biol. 2018;80:50–64. doi:10.1016/j.semcdb.2017.05.023
82. Hayes JD, McLellan LI. Glutathione and glutathione-dependent enzymes represent a co-ordinately regulated defence against oxidative stress. Free Radic Res. 1999;31(4):273–300. doi:10.1080/10715769900300851
83. Strange RC, Spiteri MA, Ramachandran S, Fryer AA. Glutathione-S-transferase family of enzymes. Mutat Res. 2001;482(12):21–26. doi:10.1016/S0027-5107(01)00206-8
84. Dusinska M, Staruchova M, Horska A, et al. Are glutathione S transferases involved in DNA damage signalling? Interactions with DNA damage and repair revealed from molecular epidemiology studies. Mutat Res. 2012;736(12):130–137. doi:10.1016/j.mrfmmm.2012.03.003
85. Lusini L, Tripodi SA, Rossi R, et al. Altered glutathione anti-oxidant metabolism during tumor progression in human renal-cell carcinoma. Int J Cancer. 2001;91(1):55–59. doi:10.1002/1097-0215(20010101)91:1<55::AID-IJC1006>3.0.CO;2-4
86. Pljesa-Ercegovac M, Mimic-Oka J, Dragicevic D, et al. Altered antioxidant capacity in human renal cell carcinoma: role of glutathione associated enzymes. Urol Oncol. 2008;26(2):175–181. doi:10.1016/j.urolonc.2007.02.007
87. Llavanera M, Delgado-Bermudez A, Olives S, et al. Glutathione S-transferases play a crucial role in mitochondrial function, plasma membrane stability and oxidative regulation of mammalian sperm. Antioxidants. 2020;9:2.
88. Yin Y, Zhu P, Luo T, Xia X. Association of single-nucleotide polymorphisms in antioxidant genes and their gene-gene interactions with risk of male infertility in a Chinese population. Biomed Rep. 2020;13(1):49–54. doi:10.3892/br.2020.1306
89. Chu G, Li Y, Dong X, Liu J, Zhao Y. Role of NSD1 in H2O2-induced GSTM3 suppression. Cell Signal. 2014;26(12):2757–2764. doi:10.1016/j.cellsig.2014.08.026
90. Liu X, Campbell MR, Pittman GS, Faulkner EC, Watson MA, Bell DA. Expression-based discovery of variation in the human glutathione S-transferase M3 promoter and functional analysis in a glioma cell line using allele-specific chromatin immunoprecipitation. Cancer Res. 2005;65(1):99–104.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.