Back to Journals » Cancer Management and Research » Volume 11
Growth and differentiation factor 15 regulates PD-L1 expression in glioblastoma
Authors Peng H, Li Z, Fu J, Zhou R
Received 24 October 2018
Accepted for publication 20 February 2019
Published 2 April 2019 Volume 2019:11 Pages 2653—2661
DOI https://doi.org/10.2147/CMAR.S192095
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Lu-Zhe Sun
Haiqin Peng, Zhanzhan Li, Jun Fu, Rongrong Zhou
Department of Oncology, Xiangya Hospital, Central South University, Changsha, People’s Republic of China
Background: Gliomablastoma multiforme (GBM) is the most fatal form of all brain cancers in human with no successful treatment available. Programmed death-ligand 1 (PD-L1) is a coinhibitory ligand predominantly expressed by tumor cells. Growth differentiation factors (GDFs) are a subfamily of proteins belonging to the transforming growth factor beta superfamily that have functions predominantly in tissue development and cancer.
Purpose: To investigat the expression of GDFs in GBMs, and explored the potential regulatory role of GDFs on PD-L1 expression in GBMs.
Methods: GEO2R program were analyzed for the mRNA expression data of GDFs in GSE4290 dataset. Analysis of TCGA GBM datasets were further determined the relationship between GDFs and PD-L1. Western blot Western blot was used to detect the expression of PD-L1 in GBM cell lines.
Results: GDFs displayed differential patterns of expression with GDF15 and myostatin (MSTN) highly enriched in GBM tissues. We also identified GDF15 as a novel regulator that induces PD-L1 expression in GBM cells. Consistently, GDF15 expression correlated with PD-L1 in TCGA GBM dataset. Further, GDF15 enhanced PD-L1 expression via Smad2/3 pathway in GBM cell line U87, U251 and SHG44, which was inhibited by Smad2/3 inhibitor SIS3. Knockdown of GDF15 attenuated Smad2/3 signaling and reduced PD-L1 expression in A172 and GIC6 glioma cells.
Conclusion: GDF15 might be a novel regulator of PD-L1 expression in GBMs; targeting GDF15/PD-L1 pathway might be a promising therapeutic approach for GBM patients.
Keywords: PD-L1, GDF, GDF15, GBM, immunotherapy
Introduction
Glioblastoma (GBM) is the most common primary malignant tumor in the central nervous system, with an incidence of approximately 3/100,000 persons per year.1,2 The standard therapy for newly diagnosed GBM involves surgical resection combined with chemotherapy and/or radiotherapy. Although advances in radiotherapy and chemotherapy have brought modest improvements in the survival of patients with malignant GBM, the invasive nature of the disease still leads to a poor prognosis, with a median overall survival of 14.9 months and a 5 year survival of only 5.5% following standard-of-care treatment.3 Thus, new treatment strategies for malignant gliomas are urgently needed.
Immunotherapy has recently emerged as the fourth pillar of cancer treatment, together with surgery, radiation, and chemotherapy. Blockade of immune checkpoints has been the most promising approach to activating antitumor immunity; of the immune checkpoints, programmed cell death 1 (PD-1) is the most effective and widely studied.4,5 The anti-PD-1 antibodies nivolumab and pembrolizumab were recently approved by the US Food and Drug Administration (FDA) for the treatment of melanoma, non-small cell lung cancer (NSCLC), and other malignant tumors.6–8 Programmed death-ligand 1 (PD-L1), the ligand for PD-1, is predominantly expressed in cancer cells.9 PD-L1 expressed in various cancer cells can help immune evasion by interacting with PD-1 on T cells.10 Recently, a new study demonstrated that intrinsic PD-L1 had prominent oncogenic effects, preferentially in aspects of promoting migration and invasion, on GBM cells in vitro and in vivo.11 In GBM, PD-L1 expression levels range from 7.8% to 37.5% in tumor-resident cells.12,13 High expression of PD-L1 in cancer cells is correlated with a poor prognosis.3 Although preliminary clinical study (CheckMate 143) demonstrated a failure of nivolumab to prolong overall survival of patients with recurrent glioma, progress in research into PD-L1 would enable us to develop a more effective and individualized immunotherapeutic strategy for GBM.14
Growth and differentiation factors (GDFs) are members of the transforming growth factor (TGF)-β superfamily. Although the important roles of GDFs in bone development, angiogenesis, embryonic development, inflammatory responses, and acute injury have been documented, there is little knowledge on the functions of GDFs in cancer.15 Varadaraj et al demonstrated that GDF2 has anti-metastatic capabilities in ovarian and breast cancers.16 GDF5 regulates TGF-β-dependent angiogenesis in breast carcinoma MCF-7 cells.17 GDF9 could negatively regulate the aggressive behavior of human bladder cancer cells.18 GDF3 inhibits the growth of breast cancer cells and promotes the apoptosis induced by Taxol.19 These studies indicate that GDFs might play an important role in tumor development. Among GDFs, the role of GDF15 in cancer has been widely investigated. GDF15 expression was associated with poor prognosis and cell proliferation in non-small cell lung cancer.20 GDF15 also promotes the proliferation of cervical cancer cells by phosphorylating AKT1 and Erk1/2 through the receptor ErbB2.21 GDF15 promotes the migration and invasion of glioma cells with malignant phenotype.22 Currently, there are few literatures regarding the immunomodulatory role of GDFs in cancer. Roth et al showed that GDF15 might participate in the local immunosuppressive environment around glioma through paracrine action, thus contributing to the proliferation and immune escape of malignant glioma.23 However, how GDF15 helps the tumor cells evade the immune surveillance in GBM still remains poorly understood.
In this study, we aimed to investigate the expression of GDFs in GBM and to explore the potential regulatory role of GDFs on PD-L1 expression. We identified GDF15 and MSTN as potential tumor-promoting genes highly expressed in GBMs. We further revealed a potential regulatory role of GDF15 in activation of PD-L1 via Smad2/3 signaling, which might implicate GDF15/PD-L1 as a potential target for immunotherapy in GBM.
Materials and methods
Reagents and cell lines
The human GBM cell lines U87, A172 and U251 were obtained from the American Type Culture Collection and cultured according to the manufacturer’s protocol. Human GBM cell lines SHG44 and TJ905 were kindly provided by the Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China. GBM cell lines were routinely cultured in Dulbecco’s Modified Eagle Medium (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (HyClone, South Logan, UT, USA) at 37 °C in humidified air with 5% CO2. Glioma initiating cell line GIC6 was established and cultured as we described previously.24 GDFs were purchased from PeproTech (Rocky Hill, NJ, USA). Antibodies against p-Smad2 (Ser465/467)/p-Smad3 (Ser423/425), and Smad2/3 were purchased from Cell Signaling Technology (Danvers, MA, USA). β-actin antibody was from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibodies against PD-L1 and GDF15 were from Abcam (Cambridge, MA, USA). Horseradish peroxidase-conjugated goat anti-rabbit and anti-mouse antibodies were purchased from Bio-Rad (Hercules, CA, USA). PI3K/AKT inhibitor LY294002, Smad2/3 inhibitor SIS3, and the MEK/ERK inhibitor PD98059 were from Selleckchem (Houston, TX, USA). Lentiviral plasmids expressing scramble or shGDF15 were purchased from Genecopoeia Inc. (Rockville, MD, USA). The packaging procedure for lentiviral shRNAs was conducted as described previously.25,26 pCMV3-C-FLAG plasmid expressing PD-L1 cDNA ORF or empty vector control was purchased from Sinobiological (Wayne, PA, USA). siRNAs targeting Smad2/3 or scramble control were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Western blot analysis
Cell lysates were prepared using cell lysis buffer containing 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitor cocktail. Western blot analysis was conducted as described previously.27,28 After standard SDS-PAGE and Western blot procedures, proteins were visualized by enhanced chemiluminescence (Abcam).
The Cancer Genome Atlas (TCGA) data analysis
mRNA expression data (RNA-Seq) of GBMs (n=152) were downloaded from the data portal of TCGA project (
Statistical analyses
Statistical analyses were carried out using SPSS 10.0 software (SPSS Inc., Chicago, IL, USA). Error bars indicate standard deviations. Student’s t-test was used to compare means between two groups. Kaplan–Meier curves representing overall survival were constructed, and differences in survival between groups were determined using the log-rank test. For all tests, the level of statistical significance was set at P<0.05. All statistical tests were two-sided.
Results
GDFs exhibited differential expression patterns in normal brain tissues and GBMs
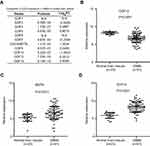
To compare the expression of GDFs in normal brain and GBM tissues, we analyzed the mRNA expression data of GDFs in GSE4290 dataset using GEO2R program. The differentially expressed GDFs in GBMs versus normal brain tissues were selected according to the following criteria: P<0.05 and |log2FC|>1. GEO2R analysis revealed differential expression patterns of GDFs in normal brain tissues (n=23) and GBMs (n=81) (Figure 1A). GBMs exhibited lower expression of GDF-10 and higher expression of GDF15 and GDF-8/MSTN than normal brain tissues (P<0.05, Figure 1B–D). These results suggested that GDF15 and MSTN might promote tumor development in GBM.
GDFs exhibited differential effects on the expression of PD-L1 in GBM cells
The expression of PD-L1 in GBMs was also analyzed based on GSE4290 dataset. As shown in Figure 2A, PD-L1 exhibited higher expression in GBMs than those in normal brain tissues (P=0.0042). Higher expression of PD-L1 was also associated with unfavorable prognosis in TCGA GBM patients (n=152, P=0.0498, Figure 2B). We next performed preliminary analysis to determine the effects of GDFs on the expression of PD-L1 in U87 cells. PD-L1 expression was significantly increased in U87 cells incubated with MSTN and GDF15, but reduced by GDF3 and GDF7 (Figure 2C and D). The other GDFs only displayed mild effect on PD-L1 expression in U87 cells.
GDF15 expression was correlated with PD-L1 in GBM cell lines and GBM patients
We analyzed TCGA GBM datasets to further confirm the relationship between GDFs and PD-L1. The mRNA expression levels of GDF15 significantly correlated with those of PD-L1 in the TCGA GBM datasets (n=152, Spearman’s correlation r=0.51, P<0.05, Table 1). In contrast, the expression of PD-L1 did not significantly correlate with that of MSTN (Table 1). Moreover, we did not observe a significant correlation between IFN-γ and GDF15 in TCGA GBM dataset (Spearman correlation r=0.0309, P=0.704). Therefore, GDF15 might be a novel regulator of PD-L1 expression in GBMs.
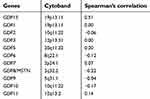 | Table 1 Correlation between PD-L1 and GDFs in GBM (TCGA RNA-seq dataset, n=152) |
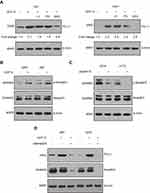
GDF15 regulated PD-L1 expression in GBM cell lines
Western blotting analysis showed that PD-L1 expression was largely consistent with GDF15 in GBM cell lines, with higher expression in A172, TJ905 and GIC6 cells (Figure 3A). To confirm the effects of GDF15 on GBM cells, two GBM cell lines (U251, SHG44) were transfected with GDF15 plasmid for 48 hrs. As shown in Figure 3B, over-expression of GDF15 increased PD-L1 expression in these two cell lines (Figure 3B). Furthermore, we attenuated GDF15 expression in A172 and GIC6 cells using shRNA lentivirus targeting GDF15. PD-L1 expression was markedly reduced by shRNA targeting GDF15 compared to non-targeting scramble control (Figure 3C).
GDF15 promoted PD-L1 expression via the Smad2/3 pathway in GBM cells
TGF-β activates Smad-dependent Smad2/3 signaling and Smad-independent signaling such as ERK1/2 and AKT.30 We assessed these components of TGF-β signaling pathways to explore the molecular mechanism by which GDF15 regulates PD-L1 expression in GBMs. We used the PI3K/AKT inhibitor LY294002, Smad2/3 inhibitor SIS3, and the MEK/ERK inhibitor PD98059 to abolish GDF15-induced PD-L1 expression. As shown in Figure 4A, SIS3, but not LY294002 and PD98059, effectively reduced PD-L1 expression induced by GDF15 in U87 and U251 cells, suggesting Smad2/3 might be involved in GDF15/PD-L1 signaling pathway in GBM cells. Nevertheless, we showed that GDF15 treatment increased p-Smad2/3 levels in U87 and U251 cells (Figure 4B). In contrast, knockdown of GDF15 attenuated p-Smad2/3 levels in A172 and GIC6 cells, as compared with non-targeting scramble control (Figure 4C). Moreover, the Smad2/3 expression in U87 and U251 cells was depleted by siRNAs in order to confirm the role of Smad2/3 in GDF15-induced PD-L1 expression. As shown in Figure 4D, knockdown of Smad2/3 expression attenuated PD-L1 levels induced by GDF15 in U87 and U251 cells. These results would confirm that the expression of PD-L1 is, at least in part, regulated by the GDF15/Smad2/3 signaling pathway in GBMs.
Discussion
Glioblastoma multiforme (GBM), the most common and aggressive primary brain tumor, has a high mortality rate despite extensive efforts to develop new treatments.1–3 Nivolumab and pembrolizumab (anti-PD-1 antibodies) have been approved by the US FDA for the treatment of melanoma, NSCLC, and other malignant tumors.6–8 A series of clinical trials on PD-1/PD-L1 antibodies for the treatment of glioma are currently in progress.31–33 PD-L1 is widely expressed on immunosuppressive T regulatory cells, tumor-associated macrophages, and other cells within the tumor microenvironment, including tumor cells.32 On the one hand, PD-L1 binds to PD-1 expressed on T, B, dendritic, and natural killer T cells to suppress anticancer immunity. On the other hand, the interaction between PD-L1 and CD80 initiates signals that inhibit T-cell function and cytokine production.33,34 PD-L1 expression is high in human malignant gliomas and significantly correlated with the glioma grade.9,13,35,36 These findings suggest that malignant glioma might be promoted by the selection of tumor cells with a high level of PD-L1, which facilitates immune evasion. Similarly, high expression of PD-L1 is significantly associated with a poor prognosis of GBM.37,38 Therefore, targeting PD-1/PD-L1 signaling pathway could be an effective immunotherapy for enhancing anti-tumor immunity and improving the prognosis of GBM patients.
In this study, we aimed to explore the role of GDFs in GBM. In our preliminary analysis, GDF15 and MSTN were identified as candidate tumor-promoting genes in GBM. GDF15 has been shown to play a crucial role in the occurrence and development of glioma. The addition of exogenous GDF-15 stimulated migration and invasiveness of GBM cells.22 GDF15 serum levels are a highly reliable predictor of disease progression.23 GDF15 levels were increased in the blood of GBM patients and in the cerebrospinal fluid, which correlated with poor patient outcomes.39 Recently, the role of GDF15 in immune escape of cancer cells is also documented. GDF15 participates in the local immunosuppressive environment around glioma through paracrine action.23 GDF15 is also implicated in the development of immunosuppression in triple-negative breast cancer.40 However, the regulatory mechanisms of GDF15 on immune evasion still remain poorly understood. Based on TCGA data analysis, we showed that GDF15 expression correlated with PD-L1 in TCGA GBMs. Moreover, GDF15 markedly increased PD-L1 expression in U87, U251 and SHG44 GBM cells, while knockdown of GDF15 reduced PD-L1 expression in A172 and GIC6 GBM cells. These results suggested that GDF15 might be a novel regulator of PD-L1 in GBM. We also noted the effect of other GDFs (MSTN, GDF3 and GDF7) on PD-L1 expression, which might await further investigation in the future.
GDF15 is an important regulator of multiple signaling pathways including PI3K/AKT, Smad2/3 and ERK1/2.21,41,42 However, how GDF15 regulates PD-L1 expression still remains unknown. Herein, we showed that GDF15 might promote PD-L1 expression via Smad2/3 signaling in GBM cells, as Smad2/3 inhibition effectively reduced PD-L1 expression induced by GDF15 in U87 and U251 cells. We also showed that knockdown of Smad2/3 expression attenuated PD-L1 levels induced by GDF15 in U87 and U251 cells. These results would confirm that the expression of PD-L1 is, at least in part, regulated by the GDF15/Smad2/3 signaling pathway in GBMs. Some reports indicated that Interferon-related signaling pathway could enhance PD-L1 expression in glioma.43,44 However, we did not observe the correlated expression between IFNγ and GDF15, suggesting GDF15 effect on PD-L1 might be independent of Interferon signaling in GBM. Therefore, our findings might reveal the functional link between GDF15 and PD-L1-mediated immune evasion of GBM cells, which may contribute to the tumor progression of GBM.
Conclusion
In conclusion, our findings indicate that PD-L1 expression is regulated by GDF15 via activation of the Smad2/3 signaling pathway in GBM. Our results would highlight the potential benefit of blocking GDF15/PD-L1 pathway for the treatment of malignant GBM.
Acknowledgment
This work was supported by National Nature Science Foundation of China (81672510).
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol. 2006;2(494–503):1–516. doi:10.1038/ncpneuro0096
2. Ostrom QT, Gittleman H, Liao P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol. 2014;16(Suppl 4):v1–63. doi:10.1093/neuonc/nou223
3. Nduom EK, Wei J, Yaghi NK, et al. PD-L1 expression and prognostic impact in glioblastoma. Neuro Oncol. 2016;18:195–205. doi:10.1093/neuonc/nov172
4. Lee YH, Martin-Orozco N, Zheng P, et al. Inhibition of the B7-H3 immune checkpoint limits tumor growth by enhancing cytotoxic lymphocyte function. Cell Res. 2017;27:1034–1045. doi:10.1038/cr.2017.90
5. Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi:10.1038/nrc3239
6. Chmielowski B. Ipilimumab: A first-in-class T-cell potentiator for metastatic melanoma. J Skin Cancer. 2013;2013:423829. doi:10.1155/2013/423829
7. Hamid O, Robert C, Daud A, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369:134–144. doi:10.1056/NEJMoa1305133
8. Wolchok JD, Kluger H, Callahan MK, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369:122–133. doi:10.1056/NEJMoa1302369
9. Driessens G, Kline J, Gajewski TF. Costimulatory and coinhibitory receptors in anti-tumor immunity. Immunol Rev. 2009;229:126–144. doi:10.1111/j.1600-065X.2009.00771.x
10. Luke JJ, Ott PA. PD-1 pathway inhibitors: the next generation of immunotherapy for advanced melanoma. Oncotarget. 2015;6:3479–3492. doi:10.18632/oncotarget.2980
11. Qiu XY, Hu DX, Chen WQ, et al. PD-L1 confers glioblastoma multiforme malignancy via Ras binding and Ras/Erk/EMT activation. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1754–1769. doi:10.1016/j.bbadis.2018.03.002
12. Wintterle S, Schreiner B, Mitsdoerffer M, et al. Expression of the B7-related molecule B7-H1 by glioma cells: a potential mechanism of immune paralysis. Cancer Res. 2003;63:7462–7467.
13. Berghoff AS, Kiesel B, Widhalm G, et al. Programmed death ligand 1 expression and tumor-infiltrating lymphocytes in glioblastoma. Neuro Oncol. 2015;17:1064–1075. doi:10.1093/neuonc/nou307
14. Filley AC, Henriquez M, Dey M. Recurrent glioma clinical trial, CheckMate-143: the game is not over yet. Oncotarget. 2017;8:91779–91794. doi:10.18632/oncotarget.21586
15. Rider CC, Mulloy B. Bone morphogenetic protein and growth differentiation factor cytokine families and their protein antagonists. Biochem J. 2010;429:1–12. doi:10.1042/BJ20100305
16. Varadaraj A, Patel P, Serrao A, et al. Epigenetic regulation of GDF2 suppresses anoikis in ovarian and breast epithelia. Neoplasia. 2015;17:826–838. doi:10.1016/j.neo.2015.11.003
17. Margheri F, Schiavone N, Papucci L, et al. GDF5 regulates TGFß-dependent angiogenesis in breast carcinoma MCF-7 cells: in vitro and in vivo control by anti-TGFß peptides. PLoS One. 2012;7:e50342. doi:10.1371/journal.pone.0050342
18. Du P, Ye L, Li H, Ruge F, Yang Y, Jiang WG. Growth differentiation factor-9 expression is inversely correlated with an aggressive behaviour in human bladder cancer cells. Int J Mol Med. 2012;29:428–434. doi:10.3892/ijmm.2011.858
19. Li Q, Ling Y, Yu L. GDF3 inhibits the growth of breast cancer cells and promotes the apoptosis induced by Taxol. J Cancer Res Clin Oncol. 2012;138:1073–1079. doi:10.1007/s00432-012-1213-3
20. Lu X, He X, Su J, et al. EZH2-mediated epigenetic suppression of GDF15 predicts a poor prognosis and regulates cell proliferation in non-small-cell lung cancer. Mol Ther Nucleic Acids. 2018;12:309–318. doi:10.1016/j.omtn.2018.05.016
21. Li S, Ma YM, Zheng PS, Zhang P. GDF15 promotes the proliferation of cervical cancer cells by phosphorylating AKT1 and Erk1/2 through the receptor ErbB2. J Exp Clin Cancer Res. 2018;37:80. doi:10.1186/s13046-018-0744-0
22. Codo P, Weller M, Kaulich K, et al. Control of glioma cell migration and invasiveness by GDF-15. Oncotarget. 2016;7:7732–7746. doi:10.18632/oncotarget.6816
23. Roth P, Junker M, Tritschler I, et al. GDF-15 contributes to proliferation and immune escape of malignant gliomas. Clin Cancer Res. 2010;16:3851–3859. doi:10.1158/1078-0432.CCR-10-0705
24. Liu Q, Zhang C, Yuan J, et al. PTK7 regulates Id1 expression in CD44-high glioma cells. Neuro Oncol. 2015;17:505–515. doi:10.1093/neuonc/nou227
25. Li Z, Fu J, Li N, Shen L. Quantitative proteome analysis identifies MAP2K6 as potential regulator of LIFR-induced radioresistance in nasopharyngeal carcinoma cells. Biochem Biophys Res Commun. 2018;505:274–281. doi:10.1016/j.bbrc.2018.09.020
26. Jun F, Hong J, Liu Q, et al. Epithelial membrane protein 3 regulates TGF-beta signaling activation in CD44-high glioblastoma. Oncotarget. 2017;8:14343–14358. doi:10.18632/oncotarget.11102
27. Li Z, Li N, Shen L, Fu J. Quantitative proteomic analysis identifies MAPK15 as a potential regulator of radioresistance in nasopharyngeal carcinoma cells. Front Oncol. 2018;8:548. doi:10.3389/fonc.2018.00548
28. Hu X, Chen M, Li Y, Wang Y, Wen S, Jun F. Overexpression of ID1 promotes tumor progression in penile squamous cell carcinoma. Oncol Rep. 2019;41:1091–1100. doi:10.3892/or.2018.6912
29. Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi:10.1016/j.ccr.2009.12.020
30. Colak S, Ten Dijke P. Targeting TGF-β signaling in cancer. Trends Cancer. 2017;3:56–71. doi:10.1016/j.trecan.2016.11.008
31. Huang J, Liu F, Liu Z, et al. Immune checkpoint in glioblastoma: promising and challenging. Front Pharmacol. 2017;8:242. doi:10.3389/fphar.2017.00242
32. Blumenthal DT, Yalon M, Vainer GW, et al. Pembrolizumab: first experience with recurrent primary central nervous system (CNS) tumors. J Neurooncol. 2016;129:453–460. doi:10.1007/s11060-016-2190-1
33. Carter T, Shaw H, Cohn-Brown D, Chester K, Mulholland P. Ipilimumab and bevacizumab in glioblastoma. Clin Oncol (R Coll Radiol). 2016;28:622–626. doi:10.1016/j.clon.2016.04.042
34. Bloch O, Crane CA, Kaur R, Safaee M, Rutkowski MJ, Parsa AT. Gliomas promote immunosuppression through induction of B7-H1 expression in tumor-associated macrophages. Clin Cancer Res. 2013;19:3165–3175. doi:10.1158/1078-0432.CCR-12-3314
35. Butte MJ, Keir ME, Phamduy TB, Sharpe AH, Freeman GJ. Programmed death-1 ligand 1 interacts specifically with the B7-1 costimulatory molecule to inhibit T cell responses. Immunity. 2007;27:111–122. doi:10.1016/j.immuni.2007.05.016
36. Garber ST, Hashimoto Y, Weathers SP, et al. Immune checkpoint blockade as a potential therapeutic target: surveying CNS malignancies. Neuro Oncol. 2016;18:1357–1366. doi:10.1093/neuonc/now132
37. Baral A, Ye HX, Jiang PC, Yao Y, Mao Y. B7-H3 and B7-H1 expression in cerebral spinal fluid and tumor tissue correlates with the malignancy grade of glioma patients. Oncol Lett. 2014;8:1195–1201. doi:10.3892/ol.2014.2268
38. Liu Y, Carlsson R, Ambjorn M, et al. PD-L1 expression by neurons nearby tumors indicates better prognosis in glioblastoma patients. J Neurosci. 2013;33:14231–14245. doi:10.1523/JNEUROSCI.5812-12.2013
39. Shnaper S, Desbaillets I, Brown DA, et al. Elevated levels of MIC-1/GDF15 in the cerebrospinal fluid of patients are associated with glioblastoma and worse outcome. Int J Cancer. 2009;125:2624–2630. doi:10.1002/ijc.24639
40. Rogers TJ, Christenson JL, Greene LI, et al. Reversal of triple-negative breast cancer EMT by miR-200c decreases tryptophan catabolism and a program of immunosuppression. Mol Cancer Res. 2019;17:30–41. doi:10.1158/1541-7786.MCR-18-0246
41. Liu H, Liu J, Si L, Guo C, Liu W, Liu Y. GDF-15 promotes mitochondrial function and proliferation in neuronal HT22 cells. J Cell Biochem. 2019. doi:10.1002/jcb.28339
42. Li YL, Chang JT, Lee LY, et al. GDF15 contributes to radioresistance and cancer stemness of head and neck cancer by regulating cellular reactive oxygen species via a SMAD-associated signaling pathway. Oncotarget. 2017;8:1508–1528. doi:10.18632/oncotarget.13649
43. Qian J, Wang C, Wang B, et al. The IFN-γ/PD-L1 axis between T cells and tumor microenvironment: hints for glioma anti-PD-1/PD-L1 therapy. J Neuroinflammation. 2018;15:290. doi:10.1186/s12974-018-1220-7
44. Silginer M, Nagy S, Happold C, Schneider H, Weller M, Roth P. Autocrine activation of the IFN signaling pathway may promote immune escape in glioblastoma. Neuro Oncol. 2017;19:1338–1349. doi:10.1093/neuonc/nox051
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.