Back to Journals » OncoTargets and Therapy » Volume 13
GRHL2 Acts as an Anti-Oncogene in Bladder Cancer by Regulating ZEB1 in Epithelial-Mesenchymal Transition (EMT) Process
Received 18 November 2019
Accepted for publication 13 February 2020
Published 25 March 2020 Volume 2020:13 Pages 2511—2522
DOI https://doi.org/10.2147/OTT.S239120
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Gaetano Romano
Jingang Shen, 1,* Xianbao Lv, 1,* Lei Zhang 2
1Department of Urology, Chengwu County People’s Hospital, Shandong 274200, People’s Republic of China; 2Department of Urology, Zoucheng People’s Hospital, Shandong 273500, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Lei Zhang
Department of Urology, Zoucheng People’s Hospital, No. 59, Qianquan Road, Zoucheng County, Jining, Shandong 273500, People’s Republic of China
Tel +86-13506385177
Email [email protected]
Purpose: GRHL2 played important roles in different cancers. In this study, we aimed to investigate the roles of GRHL2 in bladder cancer.
Methods: The immunohistochemistry assay was performed to detect the expression of GRHL2 in bladder cancer tissues and adjacent noncancerous tissues and the expression levels of GRHL2 and zinc finger E-box binding homeobox (ZEB1) mRNA in tissues were determined by qRT-PCR. In addition, qRT-PCR and Western blotting were applied to detect the expression levels of GRHL2 and ZEB1 in bladder cancer cell lines (RT4, BIU-87, 5637, T24) and immortalized human bladder epithelial cell line (SV-HUC-1). The cell models with up-regulated and down-regulated expression of GRHL2 were constructed using bladder cancer cell lines T24 and 5637 to investigate the underlying roles of GRHL2 on the proliferation, migration, invasion and EMT process of bladder cancer cells. After that, cell proliferation was evaluated by CCK8 assay, cell cycle assay and colony formation assay. Transwell assay and wound healing assay were performed to determine the invasion and migration ability of the bladder cancer cells. The expressions of epithelial-mesenchymal transition (EMT) related proteins (E-cadherin, Vimentin, Slug and Snail) were assessed by Western blot analysis. Moreover, ZEB1 and GRHL2 were co-transfected into T24 and 5637 cells and their effects on EMT process and invasive capacity of cells were examined.
Results: The expression of GRHL2 was down-regulated in bladder cancer tissues and human bladder cancer cell lines compared with the normal bladder tissues and immortalized human bladder epithelial cell line. Besides, down-regulation of GRHL2 improved the proliferation ability of bladder cancer cells and promoted the EMT process through up-regulation of ZEB1. The overexpression of ZEB1 partially reversed the inhibitory effect of GRHL2 on EMT.
Conclusion: GRHL2 acts as an anti-oncogene to regulate bladder cancer cell proliferation and inhibit EMT by targeting ZEB1. This study may provide a theoretical basis for further research.
Keywords: bladder cancer, GRHL2, EMT, ZEB1
Erratum for this paper has been published
Introduction
Bladder cancer has been reported as the ninth most frequent form of cancer worldwide with estimated 2.7 million people currently suffering from it.1 Bladder cancer is highly varied and the two major types of bladder cancer are muscle-invasive and non-muscle invasive bladder cancers. Moreover, a latest survey has shown that nearly 75% patients suffer from non-muscle invasive tumor whereas 15–20% of patients progress to muscle invasive tumor despite having the intravesical therapy.
Tumor metastasis includes a series of cascade reactions. Firstly, the tumor cells dispatch from the primary tumor tissue and then invaded into surrounding tissues. After that, the tumor cells enter the blood vessels for dissemination and finally extravasate into other organs.2 The initial of metastasis is the process of epithelial-mesenchymal transition (EMT), which regulates the transformation of polarized epithelial cells to highly aggressive and transitive mesenchymal cells for preparation of cell migration and invasion.3
In human, there are three Grainyhead homolog genes have been identified, known as Grainyhead-like 1–3 (GRHL1-GRHL3).4 As an epithelium-specific mammalian homolog of Drosophila Grainyhead, GRHL2 expresses in epithelial cells and regulates epidermal development.5,6 It has been suggested that GRHL2 can regulate EMT and its regulatory roles varies in different types of cancers. For example, in breast cancer, overexpressed GRHL2 has been reported to induce resistance against apoptosis through regulating death receptor ligands.7 In contrast, GRHL2 is suggested to act as a tumor suppressor in gastric and colorectal cancer cells.8,9 The debatable results indicate that GRHL2 might have tumor-specific roles in dependence of different types of cancer through regulating different targets.10 Besides that, the zinc finger E-box binding homeobox 1 and 2 (ZEB1 and ZEB2), which act as transcription factors, are also pivotal in activation of EMT.11 Both of them are known as the major target genes of GRHL2.12,13 In addition, it has been proved that GRHL2 inhibits the expression of ZEB1 by directly binding to ZEB1 promoter.
Although many evidences have proved that GRHL2 plays an important role in proliferation and metastasis of tumor,14,15 how it affects EMT in bladder cancer cells remains unclear. In this study, we aimed to find the underlying regulatory mechanism of GRHL2 in bladder cancer cells and to verify the roles of GRHL2 in EMT process by targeting ZEB1.
Materials and Methods
Clinical Specimens
Fresh bladder cancer tissues were collected from 60 patients (48 males and 12 females, 42–75 years old) hospitalized in the department of urology of Zoucheng People’s Hospital from June 2017 to January 2019. The patients were newly diagnosed with bladder cancer and had not received any chemoradiotherapy before surgery and tissue collection. Non-tumor tissues from each patient were obtained not less than 3 cm away from the edge of tumor tissue and served as the control group. The pathological specimens were classified according to the TNM criteria of Union for International Cancer Control (UICC) unites. There are 21 cases in stage T1, 15 cases in stage T2, 14 cases in stage T3, and 10 cases in stage T4, respectively. Consent was signed and obtained from all participating patients and the methodologies of this study was approved by the Ethics Committee of Zoucheng People’s Hospital and followed the Declaration of Helsinki.
Immunohistochemistry
The expression of GRHL2 in bladder tissues was evaluated by immunohistochemistry assay. In brief, the fresh bladder cancer tissues and the adjacent normal tissues were fixed in 10% natural formalin, embedded in paraffin and sliced into sections of 5 μm in thickness. After dewaxing and antigen retrieval, the paraffin-embedded tissue sections were incubated with rabbit anti-Human GRHL2 primary antibody (1:1000, abcam, Cambridge, MA, USA) at 4°C overnight according to manufacturer’s protocols. Sections were then washed and incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:200, abcam, Cambridge, MA, USA) for 2 h at room temperature. Diaminobenzidine (DAB) plus kit (TIANGEN, Beijing, China) was used for visualization. Hematoxylin (BOSTER, Wuhan, China) was then applied for counter-staining. The intensity of DAB immunostaining was quantified using Image J software (NIH, Bethesda, MD).
Cell Culture and Treatment
Human immortalized bladder epithelial cell line (SV-HUC-1) and human bladder cancer cell lines (RT4, BIU-87, 5637, T24) were purchased from Bangjing industrial co. LTD (Shanghai, China) and Cell Resource Center of the Shanghai Institutes for Biological Sciences of the Chinese Academy of Sciences (Shanghai, China), respectively. All these cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 media (Gibco, New York, USA) containing heat-inactivated fetal bovine serum (FBS, 10%, Gibco, New York, USA) at 37°C, 95%RH with 5% CO2.
Western Blot Analysis
The total proteins were firstly extracted by 100 μL lysis buffer (pierce, Rockford, IL). After that, 20 μL of protein sample was added into each column and then isolated by SDS-PAGE (Biomed, Beijing, China). Proteins were then transferred to the polyvinylidene fluoride membranes and blocked by 5% skimmed milk in TBST (BD, New Jersey, USA). The membranes were then incubated with primary antibodies against GRHL2 (GeneTex, 1:500); ZEB1 (Proteintech, 1:500); E-cadherin (Beyotime, 1:1000); Vimentin (Beyotime, 1:500); Snail (ab53519, Abcam, 1:1000); Slug (ab180714, Abcam, 1:500); GAPDH (Beyotime, 1:1000) at 4°C overnight. Then the horseradish peroxidase labeled secondary antibodies were applied with incubation for 1 h at room temperature the next day. Band intensities were normalized to GAPDH and analyzed by Image J software (NIH, Bethesda, MD).
Quantitative RT-PCR
Quantitative RT-PCR was performed to analyze the expression level of GRHL2 and ZEB1 in different cell lines. Firstly, Trizol reagent (Invitrogen, Shanghai, China) was used to extract total RNAs. The extracted RNAs were then reverse transcribed into cDNA. Quantitative RT-PCR was conducted according to the manufacturer’s instructions. Primers of GRHL2 and ZEB1 were listed as follows: GRHL2 forward primer: 5ʹ-GGAAATCTAGCCCTGGGTTTG-3ʹ, reverse primer: 5ʹ- TCAGGGAGGAACGCACTGA-3ʹ. ZEB1 forward primer: 5ʹ-ACTCTGATTCTACACCGC-3ʹ, reverse primer: 5ʹ- TGTCACATTGATAGGGCTT-3ʹ. To standardize the expression of GRHL2, GAPDH was used as an internal control and amplified by qRT-PCR with the following primers: forward primer: 5ʹ- AAGGTGAAGGTCGGAGTCAAC-3ʹ, reverse primer: 5ʹ-GGGGTCATTGATGGCAACAATA-3ʹ. The comparative CT method was used for determination of relative expression levels.
Knockdown of GRHL2
Two small interfering RNAs (siRNAs, siRNA1 and siRNA2, Genepharma, Shanghai, China) were used for knockdown of GRHL2 with the mock vehicle serving as the control group (control siRNA, Genepharma, Shanghai, China). They were transfected into T24 and 5637 bladder cancer cells by Lipofectamine 2000 (Invitrogen, California, USA) based on the manufacturer’s instructions. 48h later, qRT-PCR and Western blotting were conducted to detect the relative expression level of GRHL2 in transfected cells.
Overexpression of GRHL2/ZEB1
For buildup of GRHL2 overexpression model, the lentiviral vector expressing GRHL2 (pLVX-GRHL2 -EGFP-3FLAG-Puro) was constructed by cloning the GRHL2 gene into the pLVX-EGFP-3FLAG-Puro vector (Genepharma, Shanghai, China) and then the vectors were transfected into T24 and 5637 bladder cancer cells. The empty vector was also transfected into the cells to serve as the control group. After 48 h incubation, the transfection efficiency was determined by Western blotting. To set up the ZEB1 overexpression model, the bladder cancer cell lines were transfected with lentiviral vector expressing ZEB1 (pLVX-GRHL2 -ERFP-3FLAG-Puro). The empty vector was also transfected into the cells to serve as the control group. To construct cells that overexpressing GRHL2 and ZEB1, the blabber cancer cells were co-transfected with lentiviral vector expressing GRHL2 (pLVX-GRHL2-EGFP-3FLAG-Puro) and ZEB1 (pLVX-GRHL2-ERFP-3FLAG-Puro). After 48 h transfection, the cells were collected for analysis.
CCK8 Assay
In this study, the Cell Count Proliferation (CCK8) assay (Glpbio, Montclair, USA) was performed to detect cell viability. Briefly, the treated cells were inoculated into a 96-well plate and incubated with 10 μL CCK-8 solution at room temperature. The absorbance at different time points (24, 48, 72, 96 h) were recorded at 450nm using a microplate reader (Bio-Tek Instruments, Thermo Fisher Scientific, USA).
Colony Formation Assay
T24 and 5637 bladder cancer cells in logarithmic phase were digested by 0.25% trypsin (Thermo Fisher Scientific) and were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, and then seeded in a culture dish preheated to 37°C (density: 1000 cells/dish) with 10 mL DMEM. After incubation at 37°C with 5% CO2 for 2–3 weeks, visible colonies were fixed with 5% paraformaldehyde (Sigma-Aldrich) and then stained by 0.1% crystal violet (Sigma-Aldrich) for 30 min. After being washed with 10% PBS three times and air-dried, the clones were observed and captured by microscopy (low magnification), and colony-formation rate was calculated subsequently. Colony-formation rate = (number of cloning/number of inoculated cells)×100%.
Flow Cytometry
The cell cycle was determined by flow cytometry. In brief, 48 h after transfection with GRHL2 siRNA or empty vector GRHL2, T24 and 5637 bladder cancer cells were seeded into the 6-well plates with the density of 5×105 cells/well. Two days later, the cells were collected and re-suspended in 10% PBS, followed by application of the cell cycle analysis kit (Beyotime, Shanghai, China) and incubation for 1 h at 37°C in the dark. Tests were performed in triplicate for each sample and the cell cycle analysis was performed by FAC Scan flow cytometry (BD Biosciences, San Jose, USA) with FlowJo software according to the instructions.
Wound Healing Assay
The cells were seeded into a 6-well plate with the density of 5×106 cells per well. After the confluent monolayer was formed in DMEM containing 10% FBS, the medium was replaced by serum-free DMEM containing Mitomycin C (5 μg/mL). After that, wounds were made by a pipette tip (10 μL) scratching through the center of the monolayer. Scratch wound healing was visualized under a bright-field microscope (TE2000, Nikon) after 24 h cultivation. Migration rate was calculated by the following formula: Migration rate (% of 0 hr) = (D0-DT)/D0×100%, of which D0 represents the distance at time beginning and DT represents the distance at 24 h.
Transwell Assay
Cell invasion ability of transfected T24 and 5637 bladder cancer cells were assessed by transwell assay. Twenty-four-well chemotaxis chamber unit with 8 µm pore was purchased from Invitrogen (Shanghai, China). In short, Cells (5000) resuspended in serum-free medium were seeded into the top chamber of the well. Each well of the bottom chamber was filled with 500 μL RPMI 1640 medium containing 10% FBS. After 24 h incubation at 37°C, the cells on the upper surface of the filter were removed by wiping with a cotton swab. The invaded cells on basal side of the filter were fixed with 4% paraformaldehyde for 5 min and stained using 0.5% crystal violet for 10 min. The invasion ability was determined by the number of migrated cells in 10 randomly selected fields.
Statistical Analysis
The results in this study were expressed as mean ± standard deviation (SD). Analysis was performed by SPSS 20.0 software. The one-way analysis of variance (ANOVA) was used to detect the differences among different groups. P < 0.05 was regarded as significant difference.
Results
Expressions of GRHL2 and ZEB1 in Bladder Cancer Tissues and Cell Lines
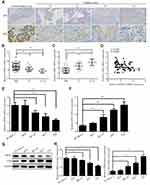
The expression level of GRHL2 in human bladder cancer tissues was analyzed by immunohistochemistry assay. It was found that GRHL2 was highly expressed in normal bladder tissues while it was down-regulated in bladder cancer tissues (Figure 1A). Furthermore, the results of qRT-PCR showed that the average mRNA level of GRHL2 was generally higher in non-tumor tissues compared with tumor tissues, meanwhile the mRNA level of GRHL2 was negatively correlated with the stage of bladder cancer progression. In details, the expression of GRHL2 mRNA was higher in stage T1+T2 than stage T3+T4 (Figure 1B) and it had close correlations with the tumor size and differentiation grade (P<0.01) (Table 1). Moreover, compared with the normal tissues, the expression of ZEB1 was higher in bladder cancer tissues, and it was negatively correlated with the expression of GRHL2 (r=−0.3297, P=0.0006) (Figure 1C and D). Additionally, the expressions of GRHL2 were significantly down-regulated at both mRNA level (Figure 1E) and protein level (Figure 1G) in RT4, BIU-87, 5637, T24 cell lines compared with SV-HUC-1 cell line, whereas the expressions of ZEB1 at both mRNA level (Figure 1F) and protein level (Figure 1H) were up-regulated in RT4, BIU-87, 5637, T24 cell lines compared with SV-HUC-1 cell line. In bladder cancer cell lines, T24 and 5637 cell lines showed much lower expression levels of GRHL2 and higher expression levels of ZEB1 compared with the normal uroepithelial cell line, thus T24 and 5637 cell lines were selected for subsequent experiments.
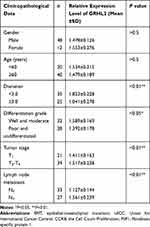 |
Table 1 Association Between the Expression Level of GRHL2 and Clinicopathological Data in Bladder Cancer Tissues |
Aberrant GRHL2 Expression Was Correlated with Cell Proliferation
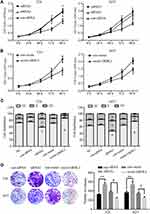
To investigate the possible effects of GRHL2 on proliferation of T24 and 5637 cells, the viability of cells transfected with GRHL2 siRNA (siRNA1 and siRNA2) or vector GRHL2 were evaluated by CCK8 assay. The cell growth curve was plotted under the observation at 450nm (Figure 2A and B). As shown in Figure 2A, the cell viability of T24 and 5637 cells transfected with GRHL2 siRNA was higher than T24 and 5637 cells transfected with the control-siRNA. On the contrary, the viabilities of T24 and 5637 cells were significantly slackened after transfected with vector-GRHL2 (Figure 2B). Because cell cycle is closely related to cellular proliferation, the effects of GRHL2 on cell cycle were further investigated. The results of flow cytometry demonstrated that in the GRHL2-knockdown cells, the average fractions of cells in S- and G2-phases were significantly increased while the fraction of cells in G1-phase was decreased. In contrast, in GRHL2 overexpressed cells, the average fraction of cells in S- and G2-phases was decreased with an increased fraction of cells in G1-phase (Figure 2C). Lastly, the colony formation assay was performed and the results showed that the cell clone formation rate was much higher in GRHL2-knockdown T24 and 5637 cells compared with the control cells. On the contrary, the cell clone formation rate was significantly decreased in GRHL2-overexpression T24 and 5637 cells (Figure 2D).
GRHL2 Regulated EMT in Bladder Cancer Cells
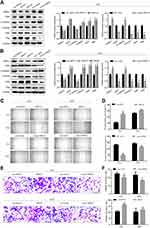
To further investigate the biological function of GRHL2, Western blotting assay was performed to assess the expression levels of EMT related proteins in GRHL2-knockdown and GRHL2-overexpression bladder cancer cell lines T24 and 5637. The results of Western blotting assay showed that ZEB1 was highly expressed in the GRHL2-knockdown cells whereas the GRHL2-overexpression cells expressed lower expression of ZEB1 compared with corresponding control groups. Furthermore, the GRHL2-knockdown cells expressed a lower level of epithelial marker (E-cadherin) and higher levels of and mesenchymal markers (Vimentin, Snail and Slug) compared with the control group (Figure 3A and B). In contrast, the GRHL2-overexpression cells exhibited a higher expression level of E-cadherin and lower levels of Vimentin, Snail and Slug in comparison with the control group (Figure 3A and B). As the results of EMT, cancer cell migration and invasion are highly related to metastasis. To further confirm the pivotal roles of GRHL2 in regulation of cell migration and invasion, wound healing assay and Transwell assay were conducted. It was found that down-regulation of GRHL2 could shorten the time of wound healing and increase the number of cells passing through the chamber compared with the cells transfected with control-siRNA (Figure 3C–F). Furthermore, the opposite effects were observed in GRHL2-overexpression cells, in which the cells needed more time for wound healing and there were fewer cells passed through the chamber compared with the cells transfected with control-vector (Figure 3C–F). Therefore, the results above clearly indicated that GRHL2 inhibited EMT process in bladder cancer cell lines T24 and 5637, it also slowed migration and invasion of bladder cancer cells.
GRHL2 Inhibited EMT Through Targeting ZEB1
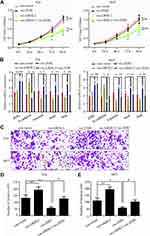
In order to explore the functional mechanism of how GRHL2 inhibited the metastasis of bladder cancer cells, the bladder cancer cells were pre-transfected with vector-GRHL2 or vector-ZEB1, respectively, or co-transfected in combination with vector-GRHL2 and vector-ZEB1. The results of CCK8 assay showed that in the T24 and 5637 cells transfected with vector-ZEB1, overexpression of ZEB1 did not change the viability of cells compared with the cells transfected with con-vector, which indicated that ZEB1 had no impact on proliferation of bladder cancer cells (Figure 4A). The results of qRT-PCR showed that overexpression of ZEB1 reduced the relative expression level of E-cadherin mRNA and promoted the relative expression levels of Vimentin, Snail and Slug mRNAs compared with the control group. Moreover, the relative expression level of E-cadherin mRNA was decreased in co-transfected cells compared with that in GRHL2-overexpression cells, but it was still higher than the control group. In contrast, the relative expression levels of Vimentin, Snail and Slug mRNAs were up-regulated in co-transfected cells compared with that in GRHL2-overexpression cells (Figure 4B), but they were still lower than the control group. Besides, similar results were obtained in Transwell assay. The bladder cancer cells co-transfected with vector-GRHL2 and vector-ZEB1 exhibited more invasive capacity compared with the cells transfected with vector-GRHL2 only (Figure 4C–E), which indicated that up-regulation of ZEB1 could partially reverse the inhibitory effect caused by GRHL2 on EMT. Altogether, it can be suggested that ZEB1 is one of the downstream targets of GRHL2 which regulates EMT but has no impact on proliferation of bladder cancer cells.
Discussion
The involvement of GRHL2 in tumorigenesis makes it become an important target in cancer therapy. To our knowledge, this is the first study to show that GRHL2 acts as a tumor suppressor gene in bladder cancer cell lines. Besides, this research provides a potential molecular evidence that overexpression of GRHL2 inhibited bladder cancer cell proliferation as well as suppressed EMT process by targeting its downstream target ZEB1. More studies are needed to well elucidate the potential roles of GRHL2 in different cancers and the knowledge can contribute to a better therapy of cancer.
Many researches have proved that GRHL2 plays a significant role in the proliferation and apoptosis of tumor cells. For instance, by inhibiting DNA methylation, GRHL2 led to an increase of hTERT expression and it enhanced cell proliferation.16 Furthermore, GRHL2 suppressed FAS (Fas cell surface death receptor) expression which induced apoptosis of cancer cells.17 Our study found that the expression level of GRHL2 was negatively correlated with the malignancy and invasiveness of bladder tumor. The expression level of GRHL2 was decreased gradually along with bladder cancer progression. Furthermore, the GRHL2 knockdown cells had higher ability to clone in vitro, while the proliferation ability was significantly reduced in GRHL2-overexpression cells. These results were consistent with the role of GRHL2 acting as a tumor suppressor gene which had been described in previous researches.16,18 However, we found that GRHL2 did not affect apoptosis, which was different from those previous studies. Collectively, these results indicated that GRHL2 acted as a tumor suppressor gene by mediating cell proliferation in bladder cancer.
At molecular level, a cell undergoing EMT needs to inhibit the expression of epithelial genes that maintains cell adhesion, to separate it from adjacent cells. E-cadherin is one of the most classical epithelial markers encoded by CDH1 gene. It is also a member of the cadherin family and constitutes a key component of adherent junctions. Several EMT transcription factors (EMY-TFs) including Snail, Slug, and ZEB1/ZEB2 directly target CDH1 for transcriptional repression. Furthermore, the EMT-TFs promote the expression of crucial mesenchymal genes such as Vimentin (VIM), fibronectin (FN1), N-cadherin (CDH2), and fibroblast-specific protein 1 (FSP1) to reshape the cytoskeleton and cell membrane to endow cells with migration ability.19,20 In this study, we observed the regulatory effect of GRHL2 on EMT by overexpressing and knocking down GRHL2. We were surprised to find that GRHL2 could regulate EMT as sensitively as a molecular switch in bladder cancer cells.
As a main target of GRHL2, ZEB1 directly binds to the promoter of E-cadherin. Yilmaz et al21 have found that the level of ZEB1 was negatively correlated with GRHL2. By inhibiting the expression of ZEB1, GRHL2 indirectly regulated the expressions of E-cadherin, Vimentin, Snail and Slug.22 Besides, during EMT, ZEB1 played an important role in improving the proliferation ability and invasiveness of tumor cells. Recent studies have found that ZEB1 is rarely expressed in epithelial tissues and highly differentiated tumor tissues, but it is highly expressed in poorly differentiated tumors, especially in the invasive part.23,24 In our study, we found that up-regulated GRHL2 directly inhibited the expression of ZEB1, indirectly inhibited the expression of E-cadherin and up-regulated the expressions of Vimentin, Snail and Slug, and vice versa. This indicated that GRHL2 was crucial in regulation of EMT. Furthermore, the inhibitory effect of GRHL2 on EMT was partially reversed in ZEB1 overexpressed cells, which indicated that ZEB1 was the downstream target for GRHL2 to exert EMT inhibition effect. However, being different from previous studies,14,18 we found that ZEB1 had no effect on the proliferation of bladder cancer cells, therefore we speculated that GRHL2 should regulate the proliferation of bladder cancer cells through other downstream signaling pathways without involvement of ZEB1. Those pathways need to be further explored and supplemented.
In conclusion, this is the first study to demonstrate the anti-tumor effect of GRHL2 in bladder cancer. The main role of GRHL2 in bladder cancer is to inhibit the EMT process by negatively regulating the expression of ZEB1. This mechanism can be partially reversed by overexpressing ZEB1.
Acknowledgment
Jingang Shen and Xianbao Lv are co-first authors for this study.
Author Contributions
Jingang Shen and Xianbao Lv performed the experiments, data analysis, collection of clinical samples and wrote the manuscript. Lei Zhang conceived and designed the study and reviewed the manuscript. All authors contributed to revising the article, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi:10.1002/ijc.29210
2. Alix-Panabières C, Mader S, Pantel K. Epithelial-mesenchymal plasticity in circulating tumor cells. J Mol Med. 2016;95(2):1–10. doi:10.1007/s00109-016-1492-2
3. Frisch SM, Schaller M, Cieply B. Mechanisms that link the oncogenic epithelial-mesenchymal transition to suppression of anoikis. J Cell Sci. 2013;126(1):21–29. doi:10.1242/jcs.120907
4. Ma L, Yan H, Zhao H, Sun J. Grainyhead-like 2 in development and cancer. Tumour Biol. 2017;39(5):101042831769837. doi:10.1177/1010428317698375
5. Werth M, Walentin K, Aue A, et al. The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development. 2010;137(22):3835. doi:10.1242/dev.055483
6. Varma S, Cao Y, Tagne J-B, et al. The transcription factors grainyhead-like 2 and NK2-homeobox 1 form a regulatory loop that coordinates lung epithelial cell morphogenesis and differentiation. J Biol Chem. 2012;287(44):37282–37295. doi:10.1074/jbc.M112.408401
7. Dompe N, Rivers CS, Li L, et al. A whole-genome RNAi screen identifies an 8q22 gene cluster that inhibits death receptor-mediated apoptosis. Proc Natl Acad Sci U S A. 2011;108(43):E943. doi:10.1073/pnas.1100132108
8. Xiang J, Fu X, Ran W, et al. Expression and role of grainyhead-like 2 in gastric cancer. Med Oncol. 2013;30(4):1–7. doi:10.1007/s12032-013-0714-5
9. Quan Y, Jin R, Huang A, et al. Downregulation of GRHL2 inhibits the proliferation of colorectal cancer cells by targeting ZEB1. Cancer Biol Ther. 2014;15(7):878–887. doi:10.4161/cbt.28877
10. Werner S, Frey S, Riethdorf S, et al. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. J Biol Chem. 2013;288(32):22993–23008. doi:10.1074/jbc.M113.456293
11. Cieply B, Farris J, Denvir J, Ford HL, Frisch SM. Epithelial-mesenchymal transition and tumor suppression are controlled by a reciprocal feedback loop between ZEB1 and Grainyhead-like-2. Cancer Res. 2013;73(20):6299–6309. doi:10.1158/0008-5472.CAN-12-4082
12. Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28(1–2):151–166. doi:10.1007/s10555-008-9179-y
13. Sanchez TE, Lazaro AR, Cuatrecasas M, et al. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29(24):3490–3500. doi:10.1038/onc.2010.102
14. Mooney SM, Talebian V, Jolly MK, et al. The GRHL2/ZEB feedback loop - a key axis in the regulation of EMT in breast cancer. J Cell Biochem. 2017;118(9):9. doi:10.1002/jcb.v118.9
15. Chen W, Yi JK, Shimane T, et al. Grainyhead-like 2 regulates epithelial plasticity and stemness in oral cancer cells. Carcinogenesis. 2016;37(5):500–510. doi:10.1093/carcin/bgw027
16. Cieply B, Farris J, Denvir J, Ford HL, Frisch SM. Epithelial-mesenchymal transition and tumor suppression are controlled by a reciprocal feedback loop between ZEB1 and Grainyhead-like-2. Cancer Res. 2013;73(20):6299–6309. doi:10.1158/0008-5472.CAN-12-4082
17. Comijn J, Berx G, Vermassen P, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7(6):1267–1278. doi:10.1016/S1097-2765(01)00260-X
18. Chung VY, Tan TZ, Tan M, et al. GRHL2-miR-200-ZEB1 maintains the epithelial status of ovarian cancer through transcriptional regulation and histone modification. Sci Rep. 2016;6(1):19943. doi:10.1038/srep19943
19. Sahay D, Leblanc R, Grunewald TG, et al. The LPA1/ZEB1/miR-21-activation pathway regulates metastasis in basal breast cancer. Oncotarget. 2015;6(24):20604–20620. doi:10.18632/oncotarget.3774
20. Ma L, Yan H, Zhao H, Sun J. Grainyhead-like 2 in development and cancer. Tumour Biol. 2017;39(5):1010428317698375. doi:10.1177/1010428317698375
21. Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28(2):15–33. doi:10.1007/s10555-008-9169-0
22. Frisch SM, Schaller M, Cieply B. Mechanisms that link the oncogenic epithelial-mesenchymal transition to suppression of anoikis. J Cell Sci. 2013;126(Pt 1):21–29. doi:10.1242/jcs.120907
23. Zhao M, Ang L, Huang J, Wang J. MicroRNAs regulate the epithelial-mesenchymal transition and influence breast cancer invasion and metastasis. Tumour Biol. 2017;39(2):1010428317691682. doi:10.1177/1010428317691682
24. Zhang J, Tian X-J, Xing J. Signal transduction pathways of EMT induced by TGF-β, SHH, and WNT and their crosstalks. J Clin Med. 2016;5(4):41. doi:10.3390/jcm5040041
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.