Back to Journals » International Journal of Nanomedicine » Volume 16
Graphene Oxide-Copper Nanocomposites Suppress Cariogenic Streptococcus mutans Biofilm Formation
Authors Mao M, Zhang W, Huang Z, Huang J, Wang J, Li W, Gu S
Received 28 January 2021
Accepted for publication 26 October 2021
Published 18 November 2021 Volume 2021:16 Pages 7727—7739
DOI https://doi.org/10.2147/IJN.S303521
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Dongwoo Khang
Mengying Mao,1– 3 Wenjie Zhang,2– 4 Zhengwei Huang,1– 3 Jing Huang,1– 3 Jia Wang,1– 3 Weiping Li,1– 3 Shensheng Gu1– 3
1Department of Endodontics, Shanghai Ninth People’s Hospital, College of Stomatology, Shanghai Jiao Tong University School of Medicine, Shanghai, People’s Republic of China; 2National Clinical Research Center for Oral Diseases, Shanghai, People’s Republic of China; 3Shanghai Key Laboratory of Stomatology & Shanghai Research Institute of Stomatology, Shanghai, People’s Republic of China; 4Department of Prosthodontics, Oral Bioengineering and Regenerative Medicine Lab, Shanghai Ninth People’s Hospital, College of Stomatology, Shanghai Jiao Tong University School of Medicine, Shanghai, People’s Republic of China
Correspondence: Shensheng Gu
Department of Endodontics, Shanghai Ninth People’s Hospital, College of Stomatology, Shanghai Jiao Tong University School of Medicine, 639 Zhizaoju Road, Shanghai, 200011, People’s Republic of China
Email [email protected]
Weiping Li
Shanghai Ninth People’s Hospital, College of Stomatology, Shanghai Jiao Tong University School of Medicine, 639 Zhizaoju Road, Shanghai, 200011, People’s Republic of China
Email [email protected]
Introduction: Dental caries is a biofilm-dependent disease that largely relies on the ability of Streptococcus mutans to synthesize exopolysaccharide matrix. Graphene oxide-based metal nanomaterials, as the derivatives of graphene, are potent agents against pathogens by their impressive antibacterial and anti-biofilm biofunctions. Previously, we fabricated the novel graphene oxide-copper nanocomposites (GO-Cu), maintaining a long-term release of copper nanoparticles. Here, the biofunctionalization of GO-Cu nanocomposites against cariogenic S. mutans is investigated.
Methods: Growth curve observation and colony forming units counting were applied to detect the antibacterial effect of GO-Cu nanocomposites on S. mutans. Scanning electron microscopy and the crystal violet assay were used to detect nanocomposite effects on biofilm forming ability. The production and distribution of exopolysaccharides within biofilm was analyzed and the expression of genes required for biofilm formation was explored. Moreover, the regulatory landscape of GO-Cu nanocomposites on S. mutans pathogenicity was probed.
Results: It has been found that GO-Gu nanocomposites were antibacterial to S. mutans and 10 μg/mL GO-Cu nanocomposites could inhibit the bacteria bioactivity instead of killing them. The biomass of S. mutans biofilm was significantly reduced when treated with 10 μg/mL GO-Cu nanocomposites. Also, 10 μg/mL GO-Cu nanocomposites could alter the biofilm architecture and impair exopolysaccharides production and distribution, and dysregulated the expression of exopolysaccharide-associated genes.
Conclusion: In all, we found low-dose GO-Cu nanocomposites could disrupt exopolysaccharide matrix assembly and further impair optimal biofilm development with minimal cytotoxicity. Therefore, GO-Cu nanocomposites can open up a new avenue for the development of alternative anti-caries biomaterials.
Keywords: graphene oxide-copper nanocomposites, anti-biofilm, Streptococcus mutans, exopolysaccharides, dental caries
Introduction
Dental caries is a typical biofilm-dependent disease, which is considered as the single most prevalent and costly oral microbial disease.1 Caries-causing biofilms consist of highly organized microbial communities that are embedded in a condensed matrix of extracellular polymeric substances.2 The matrix provides a multifunctional scaffold for structured organization and stability of the cariogenic biofilm.3,4 The major matrix components in oral biofilms associated with dental caries are exopolysaccharides, particularly Streptococcus mutans-derived glucans.5 The cariogenic S. mutans specifically metabolizes dietary sucrose into insoluble exopolysaccharides, which enhance bacterial adhesion–cohesion and accumulation on the tooth surface, promoting the formation of caries-causing acidogenic biofilm.6,7 Thus, S. mutans has been considered as a key modulator in the caries development due to its crucial capacity to synthesize and distribute exopolysaccharides in dental biofilms. Currently, more attention is focused on the development of suitable materials targeting S. mutans and, thus, to control the pathologic condition. However, the widely explored biocides, such as chlorhexidine, with strong bactericidal ability can affect the homeostasis of the oral microbiome.8 Novel approaches against S. mutans with appropriate inhibition of cell growth but enhanced efficacy inactivating exopolysaccharides production would be highly desirable.
Development of effective antibacterial material faces many challenges; it should provide effective antibacterial activity and exopolysaccharide matrix inhibition in situ, superior biocompatibility, and property of developing no resistance.9 Nowadays, ever-growing attention has been focused on metal nanoparticles due to their high reactivity and little antibiotic resistance.10,11 Copper (Cu) is an essential trace element for our health as it is a cofactor of various enzymes, but free copper is toxic to living cells.12 Copper nanoparticles (CuNPs) have been used in many fields for its multi-functional bioactivity.13,14 It is noteworthy that CuNPs can degrade the genomic DNA to exert their antibacterial effect.15 Compromising DNA in this way has a role in preventing resistance mutations and inhibiting the potential transmission of antibiotic resistance genes.16 For clinical applications, CuNPs require coatings as uncoated nanoparticles lack stability in physiological media due to their strong tendency to self-aggregation.13 Ideally, the presence of coatings would improve biofilm targeting and maintain activity, while enhancing biocompatibility, which could result in a more practical and specific anti-biofilm treatment. Graphene oxide (GO) is a product of the oxidation and exfoliation of graphite.17,18 Compared with graphene, GO has more oxygen-containing functional groups, which advantageously allows for combination with other bioactive molecules.19 In prior study, we fabricated graphene oxide-copper nanocomposites (GO-Cu), which could effectively disperse CuNPs in water by acting as a substrate to anchor nanoparticles. Also, GO-Cu nanocomposites achieved the prolonged release of CuNPs, thus enhancing the biofunctions of CuNPs.20 Based on that, we proposed GO-Cu nanocomposites with great potential as antibacterial material could target S. mutans growth and pathogenicity.
In this study, we evaluated the antibacterial and anti-biofilm properties of GO-Cu nanocomposites on cariogenic S. mutans. The effect of GO-Cu nanocomposites on exopolysaccharides matrix synthesis and exopolysaccharide-related gene expression were also detected. Moreover, the regulatory landscape of GO-Cu nanocomposites on S. mutans pathogenicity was probed. Together, we aim to propose the original use of a novel hybrid nanoparticle, the GO-Cu nanocomposites, as promising nanomaterial against cariogenic S. mutans. Exploitation of antibacterial nanomaterials can open up a new avenue for development of cariostatic approach.
Materials and Methods
Materials, Bacterial Strains and Culture Conditions
GO-Cu nanocomposites were fabricated according to the previous reports.20 The CuSO4 solution was gradually added into prepared GO solution and the final concentration of GO-Cu nanocomposites solution was defined as 1.5 mg/mL. The S. mutans wild type (WT) strain UA159 was applied as control group. The oligonucleotide primers are presented in Table 1. All primers designed using MacVector 7.0 software were purchased commercially (Sangon Biotech, Shanghai, China) and were based on the sequence of the S. mutans UA159 (ATCC 700610) genome.21 The strains were grown in brain heart infusion (BHI) medium (Oxoid, Basingstoke, UK) in an atmosphere of 80% N2 and 20% CO2 at 37°C. When needed, the medium was supplemented with 1% sucrose (BHIS).
 |
Table 1 List of Oligonucleotide Primers Used in This Study |
Growth Curve Determination
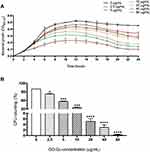
Overnight cultures of WT strains grown in BHI broth were diluted by 1:20 in fresh BHI broth and incubated at 37°C until the OD600 nm reached 0.3. Then, the diluted bacterial suspension was diluted by 1:10 with BHI broth, and was used for growth curve analysis.22 GO-Cu nanocomposites solution was diluted with culture medium at 2.5 μg/mL as the starting concentration. The 96-well plate was sealed and the bacteria were cultured at 37°C. Bacterial cell growth was monitored with a Multiskan Spectrum (SpectraMax M5, Sunnyvale, USA), and the OD600 nm were measured at 1-hour intervals. All measurements and comparisons were performed in triplicate. The representative growth curve of three independent experiments was plotted in Figure 1A.
Colony Forming Units Counting Method
Overnight cultures of WT strains grown in BHI broth were diluted by 1:20 in fresh BHI broth and cultivated to OD600 nm = 0.3. Different concentrations of GO-Cu nanocomposites solution were added into the culture and co-incubated for 2 hours.23 Bacteria treated with isotonic saline were used as control. Samples taken from each group were treated by Vortex 3 (IKA, Staufen, Germany) for cell dispersal and serially diluted, then plated onto Petri dishes. After 24-hour cultivation, the numbers of viable cells in each group were calculated by counting colony forming units (CFU).
Cell Culture, Live/Dead Assay and Cell Counting Kit-8 Assay
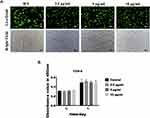
Human fibroblasts were purchased from Cyagen Bioscience (HXXFB-00001, Cyagen Bioscience, Suzhou, China) and cultured with human fibroblasts culture medium kit (HXXFB-90011) at 37°C in a humidified atmosphere with 5% CO2. Human fibroblasts were seeded in 12-well plate at a density of 10×104 cells/mL and cultured with different concentration of GO-Cu (0, 2.5, 5, 10 μg/mL). After culturing for 24 hours, the cells were washed with phosphate-buffered saline (PBS) and Calcein-AM/propidium iodide (PI) working solution (Beyotime, Shanghai, China) was added and incubated for 30 minutes at 37°C. Then the images were taken using a fluorescence microscope (Olympus, Shinjuku, Japan). Human fibroblasts were seeded 1×104 cells/well in 96-well plate. CCK-8 kit (Dojindo, Shanghai, China) was used to access the proliferation of human fibroblasts cultured with GO-Cu at day 3 and day 7. At the relevant time points, cells were washed by PBS and replaced with 100 μL CCK-8 working (serum-free human fibroblasts culture medium/CCK-8 = 10:1). After incubated in 37°C for 1 hour, the solution was transferred to a new 96-well plate and the absorbance at 450 nm was measured with a Multiskan Spectrum (SpectraMax M5, Sunnyvale, USA).
Crystal Violet Microtiter Assay
Biofilm growth was assessed as previously described.24 The biomass of the S. mutans biofilms was determined by crystal violet (CV) microtiter assay. Each well of a 96-well plate was inoculated with 2 μL exponential cultures (OD600 nm = 0.3) of bacteria and 200 μL BHIS and different concentrations of GO-Cu nanocomposites solution. After anaerobic incubation for 24 hours, the formed biofilms were washed 3 times with phosphate-buffered saline (pH 7.2) to remove non-adherent cells, air dried, and stained with 0.1% (w/v) CV for 15 mins. The bound dye was extracted from the stained cells with 1 mL destaining solution (ethanol:acetone 8:2). Biofilm formation was then quantified by measuring the optical density of the solution at 575 nm.
Confocal Laser Scanning Microscopy
Each well of a 24-well plate contained a round glass slide (diameter = 8 mm) with 20 μL exponential cultures (OD600 nm = 0.3) of bacteria and 2 mL BHIS. GO-Cu nanocomposites solution was diluted with culture medium to 10 μg/mL. To analyze the structure and viability of the nanocomposite-treated and WT S. mutans biofilms, we used confocal laser scanning microscopy (CLSM) for live/dead staining. The biofilms were stained after 24 hours of growth using the LIVE/DEAD BacLight Kit (ThermoFisher, Eugene, USA). Live bacteria were stained with SYTO 9, whereas dead bacteria were stained with propidium iodide. Biofilm images were captured by setting gates at 495–515 nm for SYTO 9 and 655–690 nm for propidium iodide. Green indicated live bacteria and red indicated dead bacteria.
For analyzing exopolysaccharide production and distribution within nanocomposite-treated and control S. mutans biofilms, we used CLSM for exopolysaccharide/bacterial staining. Briefly, 1 μM Alexa Fluor 647 (Invitrogen, Eugene, OR, USA) and 2.5 μM SYTO 9 (Invitrogen, Carlsbad, USA) were used to label exopolysaccharides and bacterial cells, respectively, in situ. The biofilms were observed by using Ar (514/488 nm) and He-Ne (543 nm) lasers. Green indicated bacteria and red indicated exopolysaccharide. A three-dimensional reconstruction of the biofilms was analyzed using Imaris 7.0.0 software (Bitplane, Zurich, Switzerland). The optical density values for the exopolysaccharides and each strain of bacteria were analyzed by ImageJ software to obtain the exopolysaccharides/bacteria optical density ratio. These experiments and calculations were repeated six times (N = 6).
Anthrone Method
Exopolysaccharides from 24 hours of 10 μg/mL GO-Cu nanocomposite-treated and WT S. mutans biofilms were extracted and purified according to previous protocols.33 For the anthrone method, standard curves were prepared with a dextran standard using various concentrations Figure S1. Four milliliters of the anthrone reagent was added to 1 mL sample solutions and heated at 95°C for 6 minutes. The absorbance of each sample at 625 nm was monitored on a microplate spectrophotometer (Shimadzu atm9700, Japan). The corresponding polysaccharide concentration was calculated according to the standard curve.
Gene Expression Assay by qRT-PCR
qRT-PCR was performed to determine the transcript levels of exopolysaccharide-associated genes in samples of nanocomposite-treated and WT 24-hour S. mutans biofilm. Each well of a 6-well plate was inoculated with 40 μL exponential cultures (OD600 nm = 0.3) of bacteria and 4 mL BHIS. GO-Cu nanocomposites solution was diluted with culture medium to 10 μg/mL. After bacterial biofilms were established for 24 hours, the biofilm samples were harvested on ice. After scraping, cells were collected by centrifugation (4500 rpm, 2422 × g) and RNA was immediately stabilized using the RNAprotect Bacteria Reagent (QIAGEN, Valencia, USA). Cells were then re-suspended in 100 mL of lysis buffer (30 mg/mL lysozyme, [pH 8.0]) and incubated at 37°C with gentle agitation for 30 minutes. RNA was extracted and purified using the classic TRIzol-chloroform protocol (Invitrogen, Carlsbad, USA). Total RNA reverse transcription was performed with the RNeasy purification kit (Qiagen, Valencia, USA). qPCR was conducted as described by the manufacturer using a Bio-Rad CFX96 TM Real-Time System (Bio-Rad, Hercules, CA, USA) and the QuantiTect SYBR-Green PCR kit (Qiagen, Valencia, USA). qPCR was performed using specific primers for the gtfB/C/D, rnc, dexA, and gbpB genes, and gyrA as a reference gene. All primers for qRT-PCR were obtained commercially (Sangon Biotech, Shanghai, China) and are listed in Table 1. We used technical replicates for each gene tested and at least 3 biological replicates in each experiment.
RNA Sequencing and Data Analysis
Samples of 10 μg/mL nanocomposite-treated and WT S. mutans were grown in fresh BHI until mid-exponential phase. Total RNA was isolated as described above. After cDNA libraries generation, the PCR products were used for sequencing by Illumina technology on Novaseq-PE150 (Illumina, San Diego, USA). Raw sequences were processed using Illumina PIPELINE software and then subjected to a series of data filtration steps for analyses. The annotated differential expression genes (DEGs) were found in GO-Cu_vs_WT comparison group. The Blast2 GO program was used to assign gene ontology (GO) terms with an E-value ≤105. Annotations of S. mutans unigenes were then used to predict biochemical pathways using the Pathway Tools. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were retrieved from KEGG web server (http://www.genome.ip/kcgg/).
Statistical Analysis
All experiments were performed in triplicate with at least 3 biological replicates. Statistical analyses of the data were performed using SPSS 16.0 (SPSS Inc., Chicago, USA). The Shapiro–Wilk and Bartlett tests were used to assess whether the data were parametric or not. For parametric testing, Fisher tests and one-way ANOVA were used to detect significant effects of variables. For nonparametric testing, the Kruskal–Wallis test and least significant difference multiple comparisons were used. Differences between means of data were considered significant if the p value was <0.05.
Results
Antibacterial Activity of GO-Cu Nanocomposites on S. mutans
In order to evaluate whether this novel GO-Cu nanomaterial could affect growth of S. mutans or not, we first pictured the growth curves of planktonic S. mutans with different concentrations of GO-Cu nanocomposites, 2.5, 5, 10, 20, 40 and 80 μg/mL, respectively. Figure 1A reveals GO-Cu nanocomposites could repress the growth of S. mutans cells. Remarkably, the repression efficiency of this nanomaterial was positively correlated to the concentration. When the concentration reached to 80 μg/mL, the growth capacity of S. mutans was seriously impaired.
Moreover, we applied a classic CFU counting method to evaluate antibacterial effect of GO-Cu nanocomposites on S. mutans viability. Similar to the results of growth curve detection, the exposure of tested bacteria to GO-Cu nanocomposites induced a relevant mortality rate in a dose-dependent manner (Figure 1B). However, we found that the CFU counting decreased sharply from 10 μg/mL group (35.1±1.56%) to 20 μg/mL group (5.1±0.98%). In summary, it has been found that GO-Gu nanocomposites were antibacterial to S. mutans and low-dose GO-Gu nanocomposites could inhibit the bacteria bioactivity instead of killing them.
The Reduced Cytotoxicity of GO-Cu Nanocomposites on Fibroblasts
As shown in Figure 2A, cells exerted strong green fluorescence and no red stained nuclei and morphological changes were seen, suggesting that human fibroblasts were viable when cultured with 2.5, 5 and 10 μg/mL of GO-Cu. Besides, CCK-8 assay was performed to detect proliferation activity of human fibroblasts. As shown in Figure 2B, after culturing with GO-Cu for 3 days and 7 days, the proliferation activity was increased and no statistically significant differences were seen between groups.
Characterization the Anti-Biofilm Effect of GO-Cu Nanocomposites
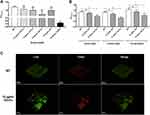
We first evaluate the anti-biofilm activity of different concentrations of GO-Gu nanocomposites on S. mutans. The biofilm of S. mutans were cultivated with 2.5, 5 and 10 μg/mL GO-Cu nanocomposites for 24 hours. The biomass of biofilms treated with 10 μg/mL significantly decreased compared to control, while other two groups treated with 2.5 and 5 μg/mL GO-Cu nanocomposites did not (Figure 3A). Thus, to investigate the specific anti-biofilm activity of GO-Cu nanocomposites, 10 μg/mL were chosen as test concentration for the subsequent experiments.
Interestingly, the biomass of 10 μg/mL GO-treated group was similar to the control group after 24-hour cultivation, indicating that the anti-biofilm activity of GO-Cu nanocomposites mainly based on the released Cu ions (Figure 3A). Besides, the dynamic biofilm formation ability of S. mutans treated with 10 μg/mL GO-Cu nanocomposites was checked. Biofilms were grown on control or GO-Cu nanocomposites, and the biomass was analyzed at various time points representing early, middle and mature-stages of development (8, 16 and 24 hours). The results showed 10 μg/mL GO-Cu nanocomposites could repress S. mutans biofilm development during all stages of biofilm formation (Figure 3B).
GO-Cu Nanocomposites Altered S. mutans Biofilm Architecture
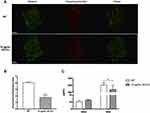
In order to compare S. mutans biofilm structure between the group with/without 10 μg/mL GO-Cu nanocomposites, confocal laser scanning microscopy (CLSM) was performed. Representative confocal images showed the WT strain formed a tenacious, well-structured and three-dimensional biofilm. On the contrary, the GO-Cu nanocomposites-treated biofilm showed small bacterial clusters sparsely distributed across the surface (Figure 3C). Similar to the CFU counting, the live/dead staining assay revealed the live bacteria (in green) in the group of GO-Cu nanocomposites slightly reduced when compared to the control group.
GO-Cu Nanocomposites Impaired Exopolysaccharides Production and Distribution
We reasoned that GO-Cu nanocomposites may impair the synthesis of exopolysaccharides, which therefore limit the formation of biofilm. To better illustrate this, we visualized the production and distribution of the glucan matrix using a dextran-conjugated cascade dye by CLSM (Figure 4). Compared to the control group, the GO-Cu nanocomposites-treated group showed less exopolysaccharides within the biofilm in two-dimensional vision. Moreover, the control group showed a three-dimensional biofilm of dense microcolonies surrounded by compact exopolysaccharides matrix. In sharp contrast, the GO-Cu nanocomposites-treated group showed dispersive exopolysaccharides and sparse cell accumulation. Specifically, the antibacterial GO-Cu nanocomposites impaired the ability of S. mutans to form exopolysaccharides, resulting in weak accumulation of bacteria and further influencing the formation of organized bacterial clusters on the surface.
GO-Cu Nanocomposites Dysregulated the Expression of Exopolysaccharide-Associated Genes
The expression profiles of the gtfB/C/D, rnc, dexA, and gbpB genes in the 24-hour biofilm phase are shown in Figure 5. Compared with the control group of WT strain without treatment in the BHIS medium, the expression of gtfB, gtfC and gbpB in 10 μg/mL GO-Cu nanocomposites-treated group was decreased, and the expression of rnc gene was up-regulated. However, the expression of gtfB, gtfC and gbpB between GO-Cu nanocomposites-treated and control group in BHI medium were similar. The data revealed that the regulation of GO-Cu nanocomposites on expression of gtfB, gtfC and gbpB genes involved in sucrose-dependent functions.
Probing the Regulatory Landscape of GO-Cu Nanocomposites on S. mutans
The RNA-seq was employed to illustrate the underlying regulatory landscape of GO-Cu nanocomposites on S. mutans pathogenicity. The samples were collected at middle-exponential phase and treated for sequencing. The Pearson correlation coefficients (PCCs) among every two samples were testified among 0.8–1, indicating the sequence data were reliable (Figure 6A). The result of RNA-seq was attached as Table S1. After a series of data analyses, the total of 26 DEGs were found in GO-Cu_vs_WT comparison group. Volcano analysis showed 5 genes were up-regulated and 21 genes were down-regulated in the 10 μg/mL GO-Cu nanocomposites-treated group, compared to the control group WT strain without treatment (Figure 6B). In particular, the expression of copZ (encoding copper chaperone, SMU_427), copY (encoding negative transcriptional regulator CopY, SMU_424) were up-regulated by 2.69-, 2.35-folds, while the expression of mreD (encoding cell shape-determining protein MreD, SMU_21) was down-regulated by 0.40-folds in GO-Cu_vs_WT comparison group (Figure 6C and D). Besides, GO analysis and KEGG pathway analysis were both applied to functional annotation of DEGs. According to GO functions, all DEGs were classified into three categories: cellular component (CC), molecular function (MF) and biological process (BP). As shown in Figure 6E, the annotated DEGs were enriched in carbohydrate transport and carbohydrate transmembrane transporter activity. Similarly, the KEGG pathway enrichment showed the fructose and mannose metabolism pathway was dominant (Figure 6F). Collectively, the annotation statistics for transcriptome analyses both showed 10 μg/mL GO-Cu nanocomposites affected carbohydrate utilization of S. mutans.
Discussion
In the present study, we provide evidence of a new hybrid nanoparticle, graphene oxide-copper nanocomposites, can effectively impair bioactivity of cariogenic S. mutans to produce exopolysaccharides and further suppress biofilm formation (Figure 7). Importantly, the antibacterial effect of GO-Cu nanocomposites at low doses (≤10 μg/mL) has been proved to mainly rely on the intrinsic dysregulation of carbohydrate metabolism, instead of direct bactericidal effect. Such properties impair glucan matrix production and thwart cariogenic biofilm accumulation, without deleterious side effects. It may lead to improved and efficacious formulations to develop potential cariostatic agents.
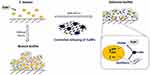 |
Figure 7 The suppression of biofilm formation of GO-Cu nanocomposites on cariogenic S. mutans. |
Unlike current antimicrobial approaches, the GO-Cu nanocomposites can exert their anti-caries ability by impairment of matrix production and slight inhibition of cell growth. Conventional antimicrobial agents mainly rely on their effective bactericidal effects. Although antibiotics have been reported to attack cariogenic pathogens efficiently, the use of antibiotics can impact negatively on the bacterial flora of oral cavity and develop resistance.10 Chlorhexidine, fluoride, hydrogen peroxide, and other chemical bactericides are capable of killing bacterial pathogens in the planktonic state, but incapable of degrading exopolysaccharides and far less effective against biofilms.25,26 Besides, the adverse effects of chlorhexidine and fluoride when usage at appropriate dosage, such as tartar formation and dental fluorosis, need to be taken into consideration.27,28 In terms of dosage, the present study confirmed the low-dose GO-Gu nanocomposites were suitable to paralyze S. mutans to form well-structured biofilms. In particular, the results of growth curves and CFU counting both estimated that the antibacterial activity of GO-Cu nanocomposites were dose-dependent, and 10 μg/mL GO-Gu nanocomposites could inhibit S. mutans growth instead of killing them. Moreover, the biomass of mature biofilm was significantly decreased in 10 μg/mL GO-Cu nanocomposites-treated group, and the scattered microcolonies core devoid of exopolysaccharides were captured by CLSM. Comparatively, the control group formed a “mushroom”-like biofilm with large amounts glucan matrix. Previous studies reported that zinc oxide29 and silver nanoparticles30 as well as iron oxide nanozymes31 have been proposed as anti-biofilm agents due to their high peroxidase-like catalytic activity in acidic environments. Compared to these antimicrobial nanomaterials, GO-Gu nanocomposites provide an important additional mechanism by repression of exopolysaccharides production for caries prevention. This application of GO-Cu nanocomposites may have broader reach as exopolysaccharides are important components in most biofilms.
Furthermore, we investigated the potential molecular mechanism underlying the repression of GO-Gu nanocomposites on exopolysaccharides production. The expression levels of exopolysaccharide-associated genes were checked at 24-hour biofilm phage. The down-regulation of gtfB and gtfC genes in 10 μg/mL GO-Cu nanocomposites-treated group was accounted for by the decreased exopolysaccharides. Briefly, the gtf gene family, known as the encoding model glucosyltransferases (Gtfs) in S. mutans, can directly synthesize exopolysaccharides from sucrose32,33. The Gtfs are responsible for glucan production that maintains the integrity of the biofilm matrix and might explain the impaired biofilms observed.7,34 However, as we all know, the genes responsible for glucan matrix formation are not only the genes directed to but also some regulator genes. In a previous study we found rnc gene could occur and function in biofilm formation, and it directly responded to stimulation and regulated its target genes by posttranscriptional repression24,35. In particular, rnc could repress the expression of vicRKX at both mRNA and protein levels. As we have known, the VicRK two-component system can promote the expression of gtfB/C genes and cell wall homeostasis.36,37 Consistent with the negative effect of rnc gene, the expression of rnc was significantly increased in 10 μg/mL GO-Cu nanocomposites-treated group. Similar to our findings, GO-AgNPs have been reported to involve in down-regulation in gene expressions of S. mutans that regulate the biological processes of adherence and quorum sensing.38 Moreover, as a complement, regulatory landscape of 10 μg/mL GO-Cu nanocomposites on S. mutans pathogenicity were probed. Among the 26 DEGs found in GO-Cu_vs_WT comparison group, the altered expressions of copZ, copY and mreD were dominant. The cariogenic pathogen S. mutans manages excess copper using the copYAZ operon that encodes a negative DNA-binding repressor (CopY), the P1-ATPase copper exporter (CopA), and the copper chaperone (CopZ).39 The increased expressions of copZ and copY confirmed that GO-Cu nanocomposites could release of CuNPs to exploit their antibacterial effect. Recent study reported in the presence of copper, transcriptional repression of gtfs was observed in copYAZ deletion strain biofilms relative to their levels of transcription in the wild-type strain biofilms.40 The negative regulatory role of copper in the transcription of the gtf genes was similar to our findings. The mreD gene has been confirmed to encode morphogenetic protein that is required for propagation of a uniform cell shape.41 The decreased expression of mreD gene indicated the nanoparticles might disrupt the homeostasis of cell shape synthesis. Besides, we enriched the annotated DEGs by GO and KEGG pathway, respectively. It is notable that both GO and KEGG analyses showed low-dose GO-Cu nanocomposites affected carbohydrate utilization of S. mutans. Collectively, low-dose GO-Cu nanocomposites could dysregulate the carbohydrate metabolism of S. mutans and led to a low amount of exopolysaccharides, which in part account for the collapsed biofilm forming ability.
Conclusion
In summary, we found GO-Cu nanocomposites could disrupt exopolysaccharide matrix assembly and further impair optimal biofilm development with minimal cytotoxicity. The nanoparticle-based therapy is indeed attractive, which could lead to a therapeutic platform to develop cariostatic approach. Future clinical studies to assess whether applications of GO-Cu nanocomposites can prevent dental caries are worthy of exploration.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China under Grant (No. 81900988 and 82071104) and Natural Science Foundation of Shanghai (No. 20ZR1448700). This work was supported by the Research Discipline fund No. KQYJXK2020 from Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, and College of Stomatology, Shanghai Jiao Tong University. This work was done in laboratory of Innovative Research Team of High-level Local Universities in Shanghai, Oral and Maxillofacial Regeneration and Functional Restoration (SSMU-ZDCX20180900).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Pitts NB, Zero DT, Marsh PD, et al. Dental caries. Nat Rev Dis Primers. 2017;3:17030. doi:10.1038/nrdp.2017.30
2. Bowen WH, Burne RA, Wu H, Koo H. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 2018;26(3):229–242. doi:10.1016/j.tim.2017.09.008
3. Kolodkin-Gal I, Cao S, Chai L. Retraction notice to: a self-produced trigger for biofilm disassembly that targets exopolysaccharide. Cell. 2015;161(4):946. doi:10.1016/j.cell.2015.04.039
4. Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14(9):563–575. doi:10.1038/nrmicro.2016.94
5. Xiao J, Klein MI, Falsetta ML, et al. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 2012;8(4):e1002623. doi:10.1371/journal.ppat.1002623
6. Koo H, Falsetta ML, Klein MI. The exopolysaccharide matrix: a virulence determinant of cariogenic biofilm. J Dent Res. 2013;92(12):1065–1073. doi:10.1177/0022034513504218
7. Klein MI, Hwang G, Santos PH, Campanella OH, Koo H. Streptococcus mutans-derived extracellular matrix in cariogenic oral biofilms. Front Cell Infect Microbiol. 2015;5:10. doi:10.3389/fcimb.2015.00010
8. Autio-Gold J. The role of chlorhexidine in caries prevention. Oper Dent. 2008;33(6):710–716. doi:10.2341/08-3
9. Garcia SS, Blackledge MS, Michalek S, et al. Targeting of Streptococcus mutans biofilms by a novel small molecule prevents dental caries and preserves the oral microbiome. J Dent Res. 2017;96:22034517698096.
10. Zanni E, Chandraiahgari CR, De Bellis G, et al. Zinc oxide nanorods-decorated graphene nanoplatelets: a promising antimicrobial agent against the cariogenic bacterium Streptococcus mutans. Nanomaterials (Basel). 2016;6:10. doi:10.3390/nano6100179
11. Liu S, Cao S, Guo J, et al. Graphene oxide-silver nanocomposites modulate biofilm formation and extracellular polymeric substance (EPS) production. Nanoscale. 2018;10(41):19603–19611. doi:10.1039/C8NR04064H
12. Rademacher C, Masepohl B. Copper-responsive gene regulation in bacteria. Microbiology. 2012;158(Pt 10):2451–2464. doi:10.1099/mic.0.058487-0
13. Wu C, Zhou Y, Xu M, et al. Copper-containing mesoporous bioactive glass scaffolds with multifunctional properties of angiogenesis capacity, osteostimulation and antibacterial activity. Biomaterials. 2013;34(2):422–433. doi:10.1016/j.biomaterials.2012.09.066
14. Lu Y, Liu X, Qiu K, et al. Facile synthesis of graphene-like copper oxide nanofilms with enhanced electrochemical and photocatalytic properties in energy and environmental applications. ACS Appl Mater Interfaces. 2015;7(18):9682–9690. doi:10.1021/acsami.5b01451
15. Warnes SL, Green SM, Michels HT, Keevil CW. Biocidal efficacy of copper alloys against pathogenic enterococci involves degradation of genomic and plasmid DNAs. Appl Environ Microbiol. 2010;76(16):5390–5401. doi:10.1128/AEM.03050-09
16. O’Gorman J, Humphreys H. Application of copper to prevent and control infection. Where are we now? J Hosp Infect. 2012;81(4):217–223. doi:10.1016/j.jhin.2012.05.009
17. Stankovich S, Dikin DA, Dommett GH, et al. Graphene-based composite materials. Nature. 2006;442(7100):282–286. doi:10.1038/nature04969
18. Zhu Y, Murali S, Cai W, et al. Graphene and graphene oxide: synthesis, properties, and applications. Adv Mater. 2010;22(35):3906–3924. doi:10.1002/adma.201001068
19. Wang Y, Li Z, Wang J, Li J, Lin Y. Graphene and graphene oxide: biofunctionalization and applications in biotechnology. Trends Biotechnol. 2011;29(5):205–212. doi:10.1016/j.tibtech.2011.01.008
20. Zhang W, Chang Q, Xu L, et al. Graphene oxide-copper nanocomposite-coated porous CaP scaffold for vascularized bone regeneration via activation of Hif-1alpha. Adv Healthc Mater. 2016;5(11):1299–1309. doi:10.1002/adhm.201500824
21. Ajdic D, McShan WM, McLaughlin RE, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci USA. 2002;99(22):14434–14439. doi:10.1073/pnas.172501299
22. Chen H, Tang Y, Weir MD, et al. Effects of S. mutans gene-modification and antibacterial monomer dimethylaminohexadecyl methacrylate on biofilm growth and acid production. Dent Mater. 2020;36(2):296–309. doi:10.1016/j.dental.2019.12.001
23. He J, Zhu X, Qi Z, et al. Killing dental pathogens using antibacterial graphene oxide. ACS Appl Mater Interfaces. 2015;7(9):5605–5611. doi:10.1021/acsami.5b01069
24. Mao MY, Li M, Lei L, Yin JX, Yang YM, Hu T. The regulator gene RNC is closely involved in biofilm formation in Streptococcus mutans. Caries Res. 2018;52(5):347–358. doi:10.1159/000486431
25. Koo H, Allan RN, Howlin RP, Stoodley P, Hall-Stoodley L. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat Rev Microbiol. 2017;15(12):740–755. doi:10.1038/nrmicro.2017.99
26. Featherstone JD, Domejean S. The role of remineralizing and anticaries agents in caries management. Adv Dent Res. 2012;24(2):28–31. doi:10.1177/0022034512452885
27. Jones CG. Chlorhexidine: is it still the gold standard? Periodontol. 1997;15:55–62. doi:10.1111/j.1600-0757.1997.tb00105.x
28. Qiu YX, Mao MY, Jiang D, Hong X, Yang YM, Hu T. Co-operative effect of exogenous dextranase and sodium fluoride on multispecies biofilms. J Dent Sci. 2015;11(1):41–47. doi:10.1016/j.jds.2015.08.001
29. Hernandez-Sierra JF, Ruiz F, Pena DC, et al. The antimicrobial sensitivity of Streptococcus mutans to nanoparticles of silver, zinc oxide, and gold. Nanomedicine. 2008;4(3):237–240. doi:10.1016/j.nano.2008.04.005
30. Ionescu AC, Brambilla E, Travan A, et al. Silver-polysaccharide antimicrobial nanocomposite coating for methacrylic surfaces reduces Streptococcus mutans biofilm formation in vitro. J Dent. 2015;43(12):1483–1490. doi:10.1016/j.jdent.2015.10.006
31. Naha PC, Liu Y, Hwang G, et al. Dextran-coated iron oxide nanoparticles as biomimetic catalysts for localized and pH-activated biofilm disruption. ACS Nano. 2019;13(5):4960–4971. doi:10.1021/acsnano.8b08702
32. Terao Y, Isoda R, Murakami J, Hamada S, Kawabata S. Molecular and biological characterization of gtf regulation-associated genes in Streptococcus mutans. Oral Microbiol Immunol. 2009;24(3):211–217. doi:10.1111/j.1399-302X.2008.00497.x
33. Lei L, Yang Y, Mao M, et al. Modulation of biofilm exopolysaccharides by the Streptococcus mutans vicX gene. Front Microbiol. 2015;6:1432. doi:10.3389/fmicb.2015.01432
34. Bowen WH, Koo H. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011;45(1):69–86. doi:10.1159/000324598
35. Mao MY, Yang YM, Li KZ, et al. The rnc gene promotes exopolysaccharide synthesis and represses the vicRKX gene expressions via MicroRNA-size small RNAs in Streptococcus mutans. Front Microbiol. 2016;7:687. doi:10.3389/fmicb.2016.00687
36. Duque C, Stipp RN, Wang B, et al. Downregulation of GbpB, a component of the VicRK regulon, affects biofilm formation and cell surface characteristics of Streptococcus mutans. Infect Immun. 2011;79(2):786–796. doi:10.1128/IAI.00725-10
37. Stipp RN, Boisvert H, Smith DJ, et al. CovR and VicRK regulate cell surface biogenesis genes required for biofilm formation in Streptococcus mutans. PLoS One. 2013;8(3):e58271. doi:10.1371/journal.pone.0058271
38. Kulshrestha S, Qayyum S, Khan AU. Antibiofilm efficacy of green synthesized graphene oxide-silver nanocomposite using Lagerstroemia speciosa floral extract: a comparative study on inhibition of gram-positive and gram-negative biofilms. Microb Pathog. 2017;103:167–177. doi:10.1016/j.micpath.2016.12.022
39. Garcia SS, Du Q, Wu H. Streptococcus mutans copper chaperone, CopZ, is critical for biofilm formation and competitiveness. Mol Oral Microbiol. 2016;31(6):515–525. doi:10.1111/omi.12150
40. Singh K, Senadheera DB, Levesque CM, Cvitkovitch DG. The copYAZ operon functions in copper efflux, biofilm formation, genetic transformation, and stress tolerance in Streptococcus mutans. J Bacteriol. 2015;197(15):2545–2557. doi:10.1128/JB.02433-14
41. White CL, Kitich A, Gober JW. Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol Microbiol. 2010;76(3):616–633. doi:10.1111/j.1365-2958.2010.07108.x
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.