Back to Journals » OncoTargets and Therapy » Volume 13
Grainyhead-Like Genes Family May Act as Novel Biomarkers in Colon Cancer
Authors Yuan M, Wang J, Fang F
Received 17 December 2019
Accepted for publication 22 February 2020
Published 17 April 2020 Volume 2020:13 Pages 3237—3245
DOI https://doi.org/10.2147/OTT.S242763
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr XuYu Yang
Minchi Yuan,1 Jianping Wang,2 Fazhuang Fang3
1Department of Oncology, The First People’s Hospital of Jiashan, Jiashan, Zhejiang, People’s Republic of China; 2Department of Anorectal Surgery, Lishui Hospital of Zhejiang University, Zhejiang, People’s Republic of China; 3Department of Abdominal Tumor Surgery, Jinhua Guangfu Hospital, Jinhua, Zhejiang, People’s Republic of China
Correspondence: Fazhuang Fang
Department of Abdominal Tumor Surgery, Jinhua Guangfu Hospital, No. 1296 Huancheng North Road, Jinhua, Zhejiang 321000, People’s Republic of China
Tel +86-18367938653
Email [email protected]
Objective: The Grainyhead-like (GRHL) genes family were reported to participate in the development of a number of diseases. This study was designed to investigate the role of GRHL genes family in colon cancer (CC).
Methods: In this study, the transcriptional levels of GRHL genes family in patients with CC from GEPIA were explored. Meanwhile, the immunohistochemical data of the GRHL genes family were also obtained in the HPA database. Additionally, we re-identified the mRNA of these genes via real-time PCR. Furthermore, the association between the levels of GRHL genes and stage plot as well as survival condition including overall survival and disease-free survival of patients with CC was analyzed. Finally, by transfecting with specific-siRNA, clone formation assay was performed to observe the role of GRHL genes family in the proliferation of SW480 human colon cancer cells.
Results: We found that the mRNA and protein levels of GRHL1, GRHL2 and GRHL3 were significantly higher in CC tissues than in normal colon tissues. Additionally, GRHL1, GRHL2 and GRHL3 were significantly associated with the stages of CC. The Kaplan–Meier plotter showed that the low levels of GRHL1, GRHL2 and GRHL3 conferred a better overall survival of patients with CC while the high levels of GRHL1 and GRHL3 were associated with poor disease-free survival. Knockdown of GRHL1, GRHL2 and GRHL3 siHgnificantly inhibited the ability of colony formation of human colon cancer cells.
Conclusion: Our study demonstrated that GRHL genes are involved in the prognosis and survival in patients with CC, the inhibition of which may suppress the proliferation of colon cancer cells.
Keywords: colon cancer, Grainyhead-like genes family, prognosis, biomarkers, bioinformatics
Introduction
Colon cancer (CC) is one of the most prevalent malignant tumors, with a high recurrence and the fourth most common cause of cancer-related deaths globally1,2. The prognosis of CC is predominantly determined by the clinicopathological features and tumor stages.3 However, the heterogeneity of the CC makes it is not easy to predict patient prognosis according to these conventional strategies.4 Additionally, the 5-year overall survival rate of CC remains still very low.5 Hence, a set of sensitive prognostic markers and potential drug targets should be identified to improve the accuracy of prognosis and individualized treatments.
The Grainyhead-like (GRHL) family of transcription factors possess 3 members, namely GRHL1, GRHL2 and GRHL3, which were firstly discovered in the fruit fly Drosophila melanogaster.6 The 3 transcription factors were found to adopt a DNA-binding immunoglobulin fold homologous to the key domain of p53 (a well-known tumor suppressor).7 The expression of the 3 transcription factors is in a tissue and developmentally specific manner, which is primarily expressed in epithelial tissues of organs including olfactory and oral epithelium, urogenital tract, kidney, digestive tract, lung and myocardium.8 At present, a great many studies unveiled the roles of GRHL genes in different types of cancer: gastric cancer, squamous cell carcinoma of the skin, breast cancer, prostate cancer, colorectal cancer, hepatocellular carcinoma, clear cell renal cell carcinoma, neuroblastoma, and cervical cancer.6,9 The roles of GRHL genes in various types of cancer are complicated, and even in some cases appear to be contradictory: sometimes they promote cancer development, sometimes they may serve as tumor suppressors. For example, knockdown of GRHL2 in colorectal cancer cells could inhibit cell proliferation by targeting ZEB1.10 Meanwhile, the protein level of GRHL2 was significantly higher in colorectal cancer tissues, which was positively correlated with tumor size and TNM stage. Furthermore, GRHL2 was also an independent prognostic index for recurrence-free survival and overall survival.11
The dysregulated expression levels of GRHL genes and their relationship with clinicopathological features and prognosis have been partly reported in human CC. To the best of our knowledge, bioinformatics analysis has yet been applied to investigate the roles of GRHL genes in human CC. Based on the analyses of thousands of gene expression or variation in copy numbers published online, we analyzed and identified the expression and different GRHL transcription factors in patients with CC in detail to investigate their expression patterns, potential functions, and distinct prognostic values of transcription factors in CC.
Materials and Methods
Ethics Statement
Our study was approved by the Academic Committee of The First People’s Hospital of Jiashan and conducted according to the principles expressed in the Declaration of Helsinki. All the datasets mentioned in this study were retrieved from the published literature, indicating that it was confirmed that all written informed consent was obtained. A total of 10 pairs of carcinoma tissues and adjacent tissues from patients diagnosed with CC were collected.
GEPIA (Gene Expression Profiling Interactive Analysis) Dataset
GEPIA (http://gepia.cancer-pku.cn/index.html) serves as a newly generated interactive web server designed by Zefang Tang, Chenwei Li, and Boxi Kang of Zhang Lab, Peking University, aiming to analyze the RNA sequencing expression data of 9736 tumors and 8587 normal samples from the GTEx projects the TCGA database in a standard processing manner. GEPIA provides customizable functions including tumor/normal differential expression analysis, profiling according to cancer types or pathological stages, patient survival analysis, similar gene detection, correlation analysis, and dimensionality reduction analysis.12 In this study, we mainly employed the boxplot to detect the mRNA expression of GRHL genes in CC and normal colon tissues.
The Stage Plot and Survival Condition of Patients with Different Levels of GRHL Genes
Similarly, we used the GEPIA database to obtain stage plot, overall survival and disease-free survival information of GRHL genes. The log-rank P value and hazard ratio (HR) with 95% confidence intervals were shown on the plot. P < 0.05 was statistically significant.
Human Protein Atlas
The Human Protein Atlas (HPA, https://www.proteinatlas.org/) is a Swedish-based program initiated in 2003 with the aim to map all human proteins in cells, tissues, and organs using the integration of various omics technologies, including antibody-based imaging, mass spectrometry-based proteomics, transcriptomics, and systems biology.13 By getting immunohistochemical data of patients with or without CC on the basis of HPA, we further verified the protein expression levels of GRHL genes.
Cell Culture and RNA Interference
SW480 human CC cells were acquired from the Cell Bank of Chinese Academy of Sciences. They were cultured in a 95% humidified air/5% CO2 atmosphere at 37°C in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Hyclone). The siRNA targeting human GRHL1 (siGRHL1, 5ʹ-CGAATCGTAAACGGTTCA-3′), GRHL2 (siGRHL2, 5ʹ-CGTAGCTAGTTTCGTAG-3′) and GRHL3 (siGRHL3, 5ʹ-CGAGCTTATGGGCCAAA-3′) and nonspecific siRNA (siNC) were purchased from GenePharma Company. siRNAs or siNC were inserted into SW480 human CC cells using Lipofectamine 4000 (Invitrogen) based on the manufacturer’s instructions. Th clone formation assays were performed 48 h after transfection.
Colony Formation Assay
The colony-forming ability of SW480 human CC cells transfected with siRNA or siNC was detected using a colony formation assay. In detail, the transfected cells (siGRHL1, siGRHL2, siGRHL3 and siNC) growing in log phase were trypsinized and seeded into six-well plates with a density of 2000 cells per well. The cells were kept in an incubator at 37°C for 7 days. Seven days later, the colonies were washed with phosphate-buffered saline (PBS), fixed with formalin (10%; Beyotime Institute of Biotechnology, Haimen, China) and stained with methyl violet. Finally, the methyl violet dye was washed off with PBS. The number of colonies was counted using a microscope (Olympus IX53; Olympus Corporation, Tokyo, Japan). Colony-inhibition rate=[1−(number of colonies in experimental groups/control group)]×100%; and colony-forming efficiency=(1−colony-inhibition rate) were calculated.
Real-Time PCR
Total RNA was extracted from cell cultures using Trizol (Invitrogen, Grand Island, NY, USA) based on the manufacturer’s protocol. Subsequently, the cDNA was amplified by a reverse transcriptional kit (Promega, Madison, WI, USA). The real-time PCR was performed using cDNA as a template and Universal PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA) by an Applied Biosystems 7900HT sequence detection system (Applied Biosystems, Foster City, CA, USA). The relative amount of mRNA was calculated and normalized using GAPDH as internal reference. The primers used in this study are presented in Table 1.
 |
Table 1 Gene-Specific Primers Used in Real-Time PCR |
Western Blot
Lysates from CC cells were immunoblotted according to protocol. Briefly, protein extracts were resolved on SDS-PAGE and transferred to PVDF membrane. After blocking with 5% non-fat dry milk, the membrane was incubated overnight at 4°C with primary antibodies: anti-GRHL1, anti-GRHL2, anti-GRHL3 or anti-GAPDH. After incubation with primary antibodies, membranes were incubated with secondary antibodies for 50 min. To confirm equal protein loading, blots with antibody against GAPDH were used. Proteins were visualized with Amersham ECL™ Western blotting system.
Statistical Analysis
The obtained data were presented as the mean ± SD (standard deviation) and assessed by the two-tailed Student’s t-test. A difference of P<0.05 was considered statistically significant.
Results
Transcriptional Levels of GRHL Genes in Patients with CC
By analyzing 349 normal colon tissues and 275 CC tissues based on GEPIA online website, we found that the mRNA levels of GRHL1, GRHL2 and GRHL3 were significantly higher in carcinoma tissues than in normal colon tissues (P<0.05), indicating that the GRHL family of transcription factors may act as potential markers in the diagnosis of CC (Figure 1A–C).
 |
Figure 1 The mRNA expression level of GRHL1, GRHL2 and GRHL3 (A) GRHL1; (B) GRHL2; (C) GRHL3. *P < 0.05 versus the tumor tissue. |
Immunohistochemical Data of GRHL Genes in Patients with CC
To further obtain the protein expression levels of GRHL genes, we obtained the immunohistochemical data of the 3 genes from HPA. Consistent with the above results based on GEPIA, the immunohistochemical data from HPA confirmed the protein levels of GRHL genes in CC (Figure 2A–C).
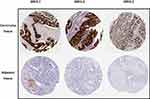 |
Figure 2 The immunohistochemical staining of GRHL genes in carcinoma tissues and adjacent tissues in patients with CC obtained from HPA database. |
Correlation Between GRHL Genes Expression and Tumor Stage in Patients with CC
Subsequently, we analyzed the association between GRHL genes expression levels and tumor stage in patients with CC based on GEPIA. The results demonstrated that the expression levels of the 3 transcription factors displayed a strong correlation with the tumor stage in patients with CC (Figure 3A–C).
 |
Figure 3 The relationship between the level of GRHL genes and tumor stages in patients with CC. (A) GRHL1; (B) GRHL2; (C) GRHL3. |
Correlation Between GRHL Genes Expression and Survival Condition in Patients with CC
Meanwhile, we further analyzed the potential association between the expression levels of GRHL genes expression levels and the survival condition of patients with CC. The Kaplan–Meier showed that the levels of 3 transcription factors displayed significant correlation with the overall survival and disease-free survival of patients with CC. In detail, the low level of 3 genes may contribute to better overall survival of CC (Figure 4A–C) while the high level of GRHL1 and GRHL3 may contribute to better disease-free survival (Figure 4D–F) (P<0.05).
 |
Figure 4 The relationship between the level of GRHL genes and overall survival (A–C) as well as disease free survival (D–F) in patients with CC; P (HR) < 0.05 versus the low GRHL TPM. |
Re-Identification of the Expression of GRHL Genes
To enhance the reliability of database, we detected the mRNA expression levels of GRHL genes in carcinoma tissues and adjacent tissues from patients with CC using real-time PCR, the results (Figure 5) of which showed further validated the previous hypothesis from bioinformatics analysis.
 |
Figure 5 Re-identification of GRHL genes expression of carcinoma tissues and adjacent tissues from patients diagnosed with CC using real-time PCR. *P < 0.05 versus the carcinoma group. |
Knockdown of GRHL Genes Reduces Proliferation of Human CC Cells
Finally, a colony formation assay and a CCK8 assay were performed to explore the effects of GRHL1, GRHL2 and GRHL3 on proliferation ability. Western blot showed that the 3 genes were successfully knocked down by siRNA Figure 6A. As shown in Figure 6B and C, the proliferation ability and the relative colony number were significantly declined after theses 3 genes were inhibited in CC cells (P<0.05). Also, we performed the same experiments to detect the proliferation ability in another cell line LoVo, the results of which further proved the anti-proliferation ability of GRHL genes (Supplementary materials).
Discussion
The dysregulation of GRHL transcription factors has been reported in various types of cancers. Although the role of GRHL transcription factors in the carcinogenesis and prognosis of some certain cancers has been partially unveiled, further bioinformatics analysis of CC on GRHL has yet to be performed till now. To our knowledge, our study is the first time to investigate the mRNA and protein expression as well as prognostic values of different members in GRHL genes family in CC. We sincerely hope that our study will contribute to available knowledge, improve treatment designs, and enhance the accuracy of prognosis for patients with CC.
The GRHL1 transcription factor is tissue-specific and is predominantly expressed in the kidney. The GRHL1 gene is located at the chromosomal position 2p25 in humans.14 In a mouse model, the loss of GRHL1 significantly affected the heart rate but had no effects on blood pressure.15 GRHL1 was also reported to serve as a tumor suppressor by affecting mild chronic skin inflammation and aberrant terminal differentiation of keratinocytes in the squamous cell carcinoma of the skin.16 Additionally, high levels of GRHL1 expression were also associated with a favorable prognosis for patients with neuroblastoma. In vitro experiments further revealed that GRHL1 could inhibit the development of neuroblastoma by regulating MYCN and HDAC3.17 In our study, we found that the mRNA and protein expression of GRHL1 was significantly higher in patients with CC, the higher level of which may be associated with tumor stage and survival condition.
GRHL2 plays vital roles in embryonic neural tube closure, epidermal integrity, and wound healing processes. Mountainous evidence has disclosed that GRHL2 may be a novel proto-oncogene which could regulate epithelial plasticity by suppressing endothelial–mesenchymal transition in several types of tumor.18 For example, GRHL2 expression level was positively associated with CD133 and E-cadherin in primary pancreatic ductal adenocarcinoma and was highly expressed in liver metastatic pancreatic ductal adenocarcinoma tissues.19 Meanwhile, Yang et al20 have identified 6 genes involving FN1, CDH2, CTNNB1, CITED2, as well as CTNNA3 as GRHL2-related genes together with GRHL2 in breast cancer metastasis, the expression levels of which were associated with clinical features. On the contrary, other studies also revealed that GRHL2 may serve as a tumor suppressor in cervical cancer, sarcoma, gastric cancer, and clear cell renal cell carcinoma.21 Exogenous GRHL2 transduced into gastric cancer cells obviously suppressed the proliferation and enhanced apoptosis. At the same time, over-expression of GRHL2 significantly reduced the protein expression levels of c-Myc and Bcl-2.22 Our study demonstrated that GRHL2 may serve as a potential proto-oncogene in CC and is involved in the proliferation of CC cells, at least proliferation ability.
GRHL3 is very important for epidermal development and homeostasis in a wide range of species.
GRHL3 deficiency could induce epidermal keratinocyte hyperproliferation during embryogenesis by targeting PTEN directly in squamous cell carcinoma.23 In colorectal cancer cells and tissues, the GRHL3 expression at both mRNA and protein levels was significantly increased. Knockdown of GRHL3 with siRNA obviously repressed colorectal cancer cell viability, proliferation, and migration, apart from promoting cell cycle arrest at G0/G1 phase and inducing cell apoptosis.24 In accordance with this study, our study also proved the expression level of GRHL3 was correlated with prognosis and tumor stage of patients with CC. More importantly, GRHL3 also affected the proliferation of CC cells, which was evidenced by the altered colony formation ability of CC cells.
Conclusion
Taken together, in this study, we systemically analyzed the expression and prognostic value of GRHL genes family in CC and provided a thorough understanding of the heterogeneity and complexity of the molecular biological properties of CC. Our studies indicated that the increased expression of GRHL1, GRHL2 and GRHL3 in CC tissues might play a vital role in CC oncogenesis; therefore, they may be promising diagnostic biomarkers for CC. Additionally, the levels of GRHL1, GRHL2 and GRHL3 were significantly associated with survival and tumor stages of the patients with CC, suggesting that Pitx1 could be potential therapeutic targets for CC.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Watanabe T, Itabashi M, Shimada Y, et al. Japanese Society for Cancer of the Colon and Rectum (JSCCR) Guidelines 2014 for treatment of colorectal cancer. Int J Clin Oncol. 2015;20(2):207–239. doi:10.1007/s10147-015-0801-z
2. Dorbeau M, Bazille C, Bibeau F. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2017;11:117–118.
3. Tesniere A, Schlemmer F, Boige V, et al. Immunogenic death of colon cancer cells treated with oxaliplatin. Oncogene. 2016;29:482–491. doi:10.1038/onc.2009.356
4. Eriksen AC, Andersen JB, Lindebjerg J, et al. Does heterogeneity matter in the estimation of tumour budding and tumour stroma ratio in colon cancer? Diagn Pathol. 2018;13:20. doi:10.1186/s13000-018-0697-9
5. Kitano S, Kitano S, Inomata M, et al. Survival outcomes following laparoscopic versus open D3 dissection for stage II or III colon cancer (JCOG0404): a Phase 3, randomised controlled trial. Lancet Gastroenterol Hepatol. 2017;2:261. doi:10.1016/S2468-1253(16)30207-2
6. Mlacki M. Recent discoveries concerning the involvement of transcription factors from the Grainyhead-like family in cancer. Exp Biol Med. 2015;240:1396–1401. doi:10.1177/1535370215588924
7. Miles LB, Dworkin S, Darido C. Alternative splicing and start sites: lessons from the Grainyhead-like family. Dev Biol. 2017;429:S0012160617303135. doi:10.1016/j.ydbio.2017.06.018
8. Ting SB, Wilanowski T, Cerruti L, et al. The identification and characterization of human Sister-of-Mammalian Grainyhead (SOM) expands the grainyhead-like family of developmental transcription factors. Biochem J. 2003;370:953–962. doi:10.1042/bj20021476
9. Werner S, Frey S, Riethdorf S, et al. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. J Biol Chem. 2013;288:22993–23008. doi:10.1074/jbc.M113.456293
10. Quan Y, Jin R, Huang A, et al. Downregulation of GRHL2 inhibits the proliferation of colorectal cancer cells by targeting ZEB1. Cancer Biol Ther. 2014;15(7):878–887. doi:10.4161/cbt.28877
11. Quan Y, Xu M, Cui P, et al. Grainyhead-like 2 promotes tumor growth and is associated with poor prognosis in colorectal cancer. J Cancer. 2015;6:342–350. doi:10.7150/jca.10969
12. Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi:10.1093/nar/gkx247
13. Uhlén M, Björling E, Agaton C, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4(12):1920–1932. doi:10.1074/mcp.M500279-MCP200
14. Liu F, Yang F, Wen D, et al. Grhl1 deficiency affects inner ear development in zebrafish. Int J Dev Biol. 2015;59:417. doi:10.1387/ijdb.140230FL
15. Pawlak M, Walkowska A, Mlącki M, et al. Consequences of the loss of the Grainyhead-like 1 gene for renal gene expression, regulation of blood pressure and heart rate in a mouse model. Acta Biochim Pol. 2015;62(2):287–296. doi:10.18388/abp.2015_1001
16. Michal M, Charbel D, Jane SM, Tomasz W. Loss of Grainy head-like 1 is associated with disruption of the epidermal barrier and squamous cell carcinoma of the skin. PLoS One. 2014;9:e89247. doi:10.1371/journal.pone.0089247
17. Fabian J, Lodrini M, Oehme I, et al. GRHL1 acts as tumor suppressor in neuroblastoma and is negatively regulated by MYCN and HDAC3. Cancer Res. 2014;74(9):2604–2616. doi:10.1158/0008-5472.CAN-13-1904
18. Tanaka Y, Kanai F, Tada M, et al. Gain of GRHL2 is associated with early recurrence of hepatocellular carcinoma. J Hepatol. 2008;49(5):746–757. doi:10.1016/j.jhep.2008.06.019
19. Gao X, Bali AS, Randell SH, Hogan BLM. GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basal stem cells. J Cell Biol. 2015;211:669–682. doi:10.1083/jcb.201506014
20. Yang X, Vasudevan P, Parekh V, Penev A, Cunningham JM. Bridging cancer biology with the clinic: relative expression of a GRHL2-mediated gene-set pair predicts breast cancer metastasis. PLoS One. 2013;8:e56195. doi:10.1371/journal.pone.0056195
21. Torresreyes LA, Alvaradoruiz L, Piñasánchez P, et al. Expression of transcription factor grainyhead-like 2 is diminished in cervical cancer. Int J Clin Exp Pathol. 2014;7:7409–7418.
22. Xiang J, Fu X, Ran W, et al. Expression and role of grainyhead-like 2 in gastric cancer. Med Oncol. 2013;30(4):1–7. doi:10.1007/s12032-013-0714-5
23. Charbel D, Georgy SR, Wilanowski T, et al. Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell. 2011;20:635–648. doi:10.1016/j.ccr.2011.10.014
24. Wang XK, Zhou FF, Tao HR, et al. Knockdown of GRHL3 inhibits activities and induces cell cycle arrest and apoptosis of human colorectal cancer cells. J Huazhong Univ Sci Technol. 2017.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

