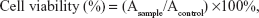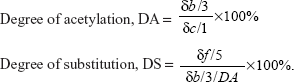Back to Journals » International Journal of Nanomedicine » Volume 9 » Issue 1
Glial cell line-derived neurotrophic factor gene delivery via a polyethylene imine grafted chitosan carrier
Authors Peng Y, Lai P, Peng S, Wu H, Yu S, Tseng T, Wang L, Chu I
Received 10 January 2014
Accepted for publication 23 March 2014
Published 28 June 2014 Volume 2014:9(1) Pages 3163—3174
DOI https://doi.org/10.2147/IJN.S60465
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Yu-Shiang Peng,1,* Po-Liang Lai,2,* Sydney Peng,1 His-Chin Wu,3 Siang Yu,1 Tsan-Yun Tseng,4 Li-Fang Wang,5 I-Ming Chu1
1Department of Chemical Engineering, National Tsing Hua University, Hsinchu, 2Department of Orthopedic Surgery, Chang Gung Memorial Hospital at Linkou, Chang Gung University College of Medicine, Taoyuan, 3Department of Materials Engineering, Tatung University, Taipei, 4Graduate School of Biotechnology and Bioengineering, College of Engineering, Yuan Ze University, Chung-Li, 5Department of Medicinal and Applied Chemistry, Kaohsiung Medical University, Kaohsiung, Taiwan
*Yu-Shiang Peng and Po-Liang Lai contributed equally to this work
Abstract: Parkinson’s disease is known to result from the loss of dopaminergic neurons. Direct intracerebral injections of high doses of recombinant glial cell line-derived neurotrophic factor (GDNF) have been shown to protect adult nigral dopaminergic neurons. Because GDNF does not cross the blood–brain barrier, intracerebral gene transfer is an ideal option. Chitosan (CHI) is a naturally derived material that has been used for gene transfer. However, the low water solubility often leads to decreased transfection efficiency. Grafting of highly water-soluble polyethylene imines (PEI) and polyethylene glycol onto polymers can increase their solubility. The purpose of this study was to design a non-viral gene carrier with improved water solubility as well as enhanced transfection efficiency for treating Parkinsonism. Two molecular weights (Mw =600 and 1,800 g/mol) of PEI were grafted onto CHI (PEI600-g-CHI and PEI1800-g-CHI, respectively) by opening the epoxide ring of ethylene glycol diglycidyl ether (EX-810). This modification resulted in a non-viral gene carrier with less cytotoxicity. The transfection efficiency of PEI600-g-CHI/deoxyribonucleic acid (DNA) polyplexes was significantly higher than either PEI1800-g-CHI/DNA or CHI/DNA polyplexes. The maximal GDNF expression of PEI600-g-CHI/DNA was at the polymer:DNA weight ratio of 10:1, which was 1.7-fold higher than the maximal GDNF expression of PEI1800-g-CHI/DNA. The low toxicity and high transfection efficiency of PEI600-g-CHI make it ideal for application to GDNF gene therapy, which has potential for the treatment of Parkinson’s disease.
Keywords: chitosan, gene transfection, glial cell, Parkinson’s disease, polyethylene imine
Introduction
Gene therapy is a promising solution to congenital and acquired diseases, such as cancer1,2 and Parkinson’s disease.3,4 The purpose of gene therapy is to provide a therapeutic gene to prevent or treat diseases related to abnormal cell function. The two main types of vectors used in gene therapy are based on either viral or non-viral gene delivery systems. Viral gene delivery systems show high transfection efficiency but exhibit disadvantages such as mutagenesis, carcinogenesis, and immune response.5–7 These safety concerns have led to the pursuit of non-viral alternatives with low immunogenicity that permit repeated administration, exhibit storage stability, and can be produced in large quantities.8,9
Chitosan (CHI) is a biodegradable, low toxic, biocompatible polysaccharide composed of β(1,4)-linked D-glucosamine and N-acetyl-D-glucosamine.10 Because of the favorable cationic property of CHI, considerable attention has been focused on CHI-based materials for use in the field of plasmid deoxyribonucleic acid (DNA) delivery, especially as non-viral vectors for gene therapy. However, the acid dissociation constant (pKa) of CHI is 6.5, which limits its solubility in acidic solutions and leads to poor gene transfection efficiency under physiological conditions.11−15
Polyethylene imine (PEI) has been shown to be a promising polycationic vector for gene delivery.16 In addition, high-molecular-weight PEI has high transfection efficiency, both in vivo and in vitro, due to its increased buffering capacity in the pH range of endosomes and lysosomes.17,18 In addition, the amine group of PEI can bind the phosphate group of DNA into nano-sized particles, protecting the DNA from degradation, shielding the DNA particles from undesired interactions, and enhancing cell binding and intercellular delivery into the cytoplasm and nucleus.19 However, the high cytotoxicity is a major concern when using PEI to deliver DNA in clinical applications.20,21
Parkinson’s disease is known to result from the loss of dopaminergic neurons in the substantia nigra. Parkinsonian symptoms are characterized by severe tremor, rigidity, bradykinesia, and postural instability.22 In particular, glial cell line-derived neurotrophic factor (GDNF) has been shown to be the most potent protective molecule for dopaminergic nigral neurons.23 GDNF has received particular attention because it can support the survival of dopaminergic midbrain neurons and therefore may provide a therapy for Parkinson’s disease. Direct intracerebral injections of high doses of recombinant GDNF have been shown to protect adult nigral dopaminergic neurons against 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine and 6-hydroxydopamine toxicity in rodents and to restore dopamine biosynthesis and tyrosine hydroxylase expression to atrophic neurons.24 Because GDNF does not cross the blood–brain barrier, intracerebral gene transfer methods are currently being investigated.25 In this work, PEI-grafted-CHI (PEI-g-CHI) was synthesized by open-ring polymerization. We screened the suitable PEI molecular weight to decrease cytotoxicity and chose the optimal DNA: polymer ratios to preserve transfection efficiency for the neuroprotective molecule GDNF.
Materials and methods
Materials
Low-molecular-weight CHI, with a viscosity of 20–300 cP, Mw 50–190 kDa, and 75%–85% deacetylation, was obtained from Aldrich (St Louis, MO, USA). PEI with an Mw of 1,800 Da (PEI1800) and poly(ethylene glycol) diglycidyl ether (EX-810) with an Mw of 512 Da were also purchased from Aldrich. PEI with an Mw of 600 Da (PEI600) was purchased from Alfa Aesar (Ward Hill, MA, USA). Dimethylsulfoxide (DMSO) and 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were supplied by Sigma (St Louis, MO, USA). The 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI), fluorescein isothiocyanate (FITC), and ethidium bromide (EtBr) were provided by Invitrogen (Camarillo, CA, USA). The propidium iodide (PI) staining solution was supplied by BD Bioscience (San Jose, CA, USA). The Dulbecco’s modified eagle medium (DMEM), antibiotic-antimycotic solution, and fetal bovine serum (FBS) were purchased from Gibco (Grand Island, NY, USA). Plasmid-encoding enhanced green fluorescence protein (GFP) (pEGFP-N1) was purchased from Clontech Laboratories, Inc., (Palo Alto, CA, USA). Plasmid-encoding GDNF and GFP (pAAC-MCS-rGDNF-IRES-hrGFP, abbreviated as pGDNF-GFP) was donated by Dr Wu at the Materials Engineering Department of Tatung University, Taiwan.
Preparation of PEI-g-CHI
Two molecular weights (600 g/mol and 1,800 g/mol) of PEI were grafted onto CHI to form PEI600-g-CHI and PEI1800-g-CHI, respectively. PEI 500 mg (Mw 600 g/mol and 1,800 g/mol, respectively) and 1.0 g of EX-810 (Mw ~440 g/mol) in conjunction with 100 mL of double deionized water were placed into a flask. The mixture was stirred at 25°C for 3 hours. Then, 100 mg of CHI was added to the mixture and titrated by 1N HCl until all the CHI was dissolved. The contents of the flask were then stirred for another 24 hours. The resultant products were CHI grafted with PEI (either PEI600-g-CHI or PEI1800-g-CHI). After reaction, the pH of the flask contents was adjusted to 9 using 1M NaOH, then transferred into two 50 mL centrifuge tubes for centrifugation at 3,000 rpm for 20 minutes. The precipitates were dissolved in an HCl solution of pH 6 then dialyzed in water under continuous flow for 3 days using a dialysis tube with a molecular weight cut-off of 6–8 kDa (Spectrum, Houston, TX, USA). The process was intended to remove the un-grafted PEI and EX-810. The product was harvested and then freeze-dried for 3 days. The dried PEI600-g-CHI and PEI1800-g-CHI were characterized by Fourier transfer infrared (FTIR) spectroscopy (Perkin–Elmer system 2000, Foster, CA, USA) and nuclear magnetic resonance (1H-NMR) spectroscopy (Unity Inova 500 NMR; Varian, Palo Alto, CA, USA) using deuterium oxide.
The endosome buffering capacity measurement
The buffering capacity of polyplex released from endosomes was measured by pH titration.26,27 The endosome buffering capability of PEI600, PEI1800, CHI, PEI600-g-CHI, and PEI1800-g-CHI were determined by acid-based titration assay over pH values ranging from high to low. The pH and pKa values of the polymers were measured by automatic titrator (702 SM Titrino; Metrohm AG, Herisau, Switzerland). A total of 500 mg of polymer was added to 150 mL of 150 mM NaCl, and the pH was adjusted to 12 using 1 M NaOH. The titration was performed at 25°C via the addition of 1 N HCl at a rate of 1 mL/min until the pH reached 1.5 under an atmosphere of CO2-free N2.
Estimation of water solubility
The solubility of CHI or PEI-g-CHI can be estimated from turbidity measurements.28,29 Briefly, 20 mg of CHI or PEI-g-CHI was dissolved in a 10 mL solution composed of 0.1 M HCl and 150 mM NaCl. The transmittance at 600 nm was measured using an ultraviolet (UV)-visible spectrophotometer (U-3300, Hitachi, Naka, Japan). The pH and transmittance of the solution were recorded after every stepwise addition of 1 M NaOH.
Amplification of plasmid DNA
Both the pEGFP-N1 plasmid and the pGDNF-GFP plasmid were introduced into Escherichia coli strain DH5α (Invitrogen, Camarillo, CA, USA). Briefly, transformed DH5α was grown in 500 mL Luria-Bertani broth containing 250 μg of antibiotics per mL and then incubated at 37°C with shaking at 200 rpm for 16 hours. Plasmid DNA was purified from DH5α by AxyPrep™ Plasmid Maxiprep Kits (Axygen Scientific Inc, Union City, CA, USA). The purity of plasmid DNA was certified by an absorbance ratio of 1.8 at OD260/OD280. The plasmid was stored at −20°C until used.
Preparation of polyplexes
PEI600-g-CHI, PEI1800-g-CHI, and CHI were each dissolved in 0.1 N HCl to obtain three 800 μg/mL polymer solutions. The polymer solutions were then diluted to various concentrations (800, 400, 200, 100, and 20 μg/mL). DNA pEGFP-N1 was dissolved in 25 mM sodium sulfate to a final concentration of 20 μg/mL. After preheating to 50°C, 250 μL DNA solutions were added dropwise into 250 μL of each of the polymer solutions under high-speed vortex for 1 minute. The DNA concentration was fixed. The polymer/DNA weight ratios were 40, 20, 10, 5, and 1. The formed polyplexes were kept at room temperature before analysis.
Characterization of polyplex
Agarose gel electrophoresis
The DNA-binding ability of CHI, PEI600-g-CHI, and PEI1800-g-CHI at various polymer/DNA weight ratios (1, 5, 10, 20, and 40) were investigated by examining the electrophoretic mobility of free DNA departing from the polyplexes on 0.8% agarose gel per 80 mL of tris-acetate-ethylenediaminetetraacetic acid (TAE) buffer (Amresco, Solon, OH, USA). Briefly, 10 μL of the DNA polyplex solution containing 0.3 μg of DNA and corresponding polymers at their respective weight ratios were mixed with 2 μL of 6X DNA-loading dye (Promega, Madison, WI, USA) and then loaded on each well. The same amount of naked DNA was used as the control. The gel was allowed to run at 100 V for 30 minutes in TAE buffer. The DNA was visualized with UV light using a FloGel FGIS-2 system (TopBio, Taipei, Taiwan).
Particle size and zeta potential
The size of the polyplexes and the surface charge were measured with a dynamic light scattering (DLS) spectroscopy (Zetasizer Nano ZS, Malvern Instruments, MA, USA) using a He-Ni laser with a wavelength of 633 nm at a scattering angle of 173°C. Approximately 1,200 μL of polyplex solution containing 5 μg of DNA was prepared at various weight ratios (1, 5, 10, 20, and 40). The mixtures were vortexed for 60 seconds before measurement. The particle size measurement was performed in triplicate at 25°C. Zeta potential measurements were performed using a capillary zeta potential cell (DTS 1061, Malvern Instruments) in automatic mode.
Transmission electron microscopy
The size and morphology of the polyplexes were observed using a transmission electron microscopy (TEM, JEM-2100, JEOL, Tokyo, Japan). Five microliters from each of the polyplexes were deposited on a carbon-coated 200-mesh copper specimen grid (Agar Scientific Ltd., Essex, UK) and allowed to dry overnight at room temperature before being examined via TEM.
Cell line experiments
Cell culture
The 293T (human embryonic kidney cell line) was used to evaluate the transfection efficiency and cytotoxicity of the polymer-DNA polyplexes. Cells were grown in DMEM supplemented with 10% FBS and 1% antibiotic-antimycotic solution. Before the experiments were performed, the cells were grown to 70%–80% confluency in an incubator at 37°C supplied with 5% CO2.
Cytotoxicity
Cytotoxicity of polymers was evaluated by MTT assay with 293T cells. PEI-25K, PEI600-g-CHI, and PEI1800-g-CHI were dissolved in fresh culture media (pH 7.4). CHI was dissolved in acidic culture medium (pH 6.0). The cells were seeded onto 96-well plates at a density of 1×104 cells/well 24 hours before the assay and then incubated at 37°C with different concentrations of polymers (5, 25, 50, 100, 250, 500, and 1,000 μg/mL) in culture media for 24 hours. A total of 50 μL of MTT reagent (5 mg/mL) was then added to each well and the cells were incubated for another 4 hours at 37°C. The media was removed and 100 μL DMSO was added to each well to dissolve the formazan crystals formed by the living cells. Cells without treatment were used as controls. Absorbance at 570 nm was recorded using a microplate reader (Synergy™ HT, BIO-Tek, Winooski, VT, USA). Cell viability was calculated according to equation 1:
| [1] |
where Asample is the absorbance of the cells treated by polymers and Acontrol is the absorbance of the untreated cells. Each experiment was performed in triplicate.
In vitro gene expression
In vitro GFP gene expression
In vitro gene transfection, using polymer/pEGFP-N1 polyplexes, was performed in 293T cells. The cells were seeded onto 24-well plates at a density of 5×104 cells/well, and cultivated in 0.5 mL of DMEM with 10% FBS. After 24 hours, the culture medium was replaced with fresh medium supplemented without serum before transfection. Polyplexes at a final volume of 100 μL with 1 μg pEGFP-N1 and various amounts of polymers (at weight ratios ranging from 1 to 40) were added to each well. After 6 hours of incubation, the culture media were replaced with 0.5 mL of fresh complete media and the cells were incubated for 72 hours post-transfection. The GFP expression was directly visualized under a fluorescence microscope (DMI 3000 B, Leica, Wetzlar, Germany).
Determination of GFP gene expression
At 72 hours after transfection, 293T cells were trypsinized for 5 minutes and then centrifuged at 1,000 rpm at room temperature for 5 minutes. The supernatant was discarded and the pellets were re-suspended with 1 mL of phosphate buffered saline (PBS) in 10% FBS. The cell expression of GFP was examined using the FACSCalibur flow cytometer (Becton Dickinson, Heidelberg, Germany) with FL1 channel set to a minimum of 1×104 cells gated per sample. Analysis was performed using Becton Dickinson CellQuest software. Data were processed using Windows Multiple Document Interface software (Scripps Research Institute, La Jolla, CA, USA).
Transfection of GDNF plasmid
In vitro gene transfection of polymer/pGDNF-GFP polyplexes was performed in 293T cells using similar methods as described for the GFP gene. The concentrations of GDNF protein in the conditioned medium were measured by enzyme-immunoassays (human GDNF DuoSet, R & D systems, Minneapolis, MN, USA). Briefly, the diluted capture antibody was incubated in 96-well plates overnight. The blocking agent was then reacted for 1.5 hours. An aliquot of supernatant was then added to the 96-well plates for another 2 hours. The recombinant human GDNF was serially diluted as a positive control in order to determine the appropriate detection range. The supernatant retrieved from the cells without transfection was used as the negative control. The biotinylated goat anti-human GDNF was added to the 96-well plates and incubated for 2 hours, and then conjugated with streptavidin-horseradish peroxidase for another 20 minutes. All reactions were performed at room temperature. Tetramethylbenzidine solution was used for staining. The reaction was stopped by the addition of 2 N sulfuric acid. The results were measured at the wavelength of 450 nm using a spectrophotometer (BioTek Synergy HT, Tustin, CA, USA).
Confocal laser scanning microscopy
The intracellular delivery of polyplexes was observed using a confocal laser scanning microscope (TCS SP II, Leica, Wetzlar, Germany). FITC-conjugated PEI600-g-CHI was synthesized by imine reaction. The 293T cells, at a density of 1×105 cells per well in 12-well plates containing one glass coverslip per well, were seeded in DMEM supplemented with 10% FBS, and incubated for 48 hours. The pEGFP-N1 plasmid was labeled with PI (1 mg/mL) at a weight ratio of 20:1 (DNA/PI). The polyplex was prepared at a PEI600-g-CHI: PI-labeled DNA weight ratio of 10:1. The 293T cells were exposed to the polyplexes in a manner identical to the transfection procedures described above and incubated at 37°C for 4 hours. After incubation, the coverslips were removed, gently washed with 2 mL of 0.1 M PBS (three times), placed in a new empty well, and treated with 1 mL of 3.7% paraformaldehyde in 0.1 M PBS for 15 minutes to fix the cells. The cells were treated with 1 mL/well of Triton X-100 and incubated for 10 minutes. After three washings with PBS, the cells were then incubated at 37°C with 0.5 mL/well of DAPI for 10 minutes. The coverslip was then mounted on the slide for imaging.
Results and discussion
Synthesis and characterization of copolymer
1H-nuclear magnetic resonance spectroscopy
We successfully synthesized PEI600-g-CHI and PEI1800-g-CHI copolymers through the reaction of epoxide rings of EX-810 with the amine groups of PEI at one end, followed by the reaction with the amine or hydroxyl groups of CHI at the other end, as shown in Figure 1. The composition of the synthesized copolymer was analyzed by 1H-NMR, as shown in Figure 2A. The peak (a) at δ =2.4–2.6 ppm is attributed to the −CH2CH2NH2 protons of PEI. The peak (b) at δ =1.9 ppm (−NHCOCH3 protons of acetyl group), the peak (c) at δ =3.1 ppm (H-2 of glucosamine ring), and the peak (d) at δ =3.4–3.81 ppm (H-3, H-4, H-5, H-6 of glucosamine ring) belong to CHI. The peak (e) at δ =3.6 ppm belongs to the −OCH2 protons of EX-810. The peak (f) at δ =4.1 ppm is attributed to methine proton after opening the epoxide rings, and was used to calculate the degree of grafting. The degree of grafting was calculated using equation 2:30
|
|
The percentages of PEI600 and PEI1800 grafted onto CHI were 16% and 19%, respectively. Although CHI is soluble only in acidic conditions, after grafting with EX-810 and PEI, PEI600-g-CHI and PEI1800-g-CHI were completely water soluble at physiological pH because of the hydrophilic nature of the added EX-810 and PEI.
FTIR spectroscopy
The absorption of [C–O–C] at 1,060 cm−1, [C–OH] at 1,019 cm−1, and [C–O] at 881 cm−1 of backbone glucosamine and acetyl-glucosamine persisted on the FTIR spectra after grafting (Figure 2B). The absorption of [CH2–CH2] at 2,928 and 2,886 cm−1 of side-chain PEI also appeared in the spectra after grafting.31 A new peak appeared at 1,065 cm−1 and was attributed to the absorption of the [C–O–C] moiety of EX-810 after linking the backbone glucosamine and the side-chain PEI. These results suggested that CHI was successfully conjugated with PEI.
Proton-buffering capacity
PEI is known to have proton-buffering capacity, resulting in disruption of endosomes in the transfection process and facilitation of DNA transport.32,33 PEI-g-CHI has a PEI moiety and, thus, may have proton-buffering capacity. The buffering capacities of PEI600-g-CHI, PEI1800-g-CHI, CHI, PEI600, and PEI1800 (using acid-base titration) are shown in Figure 3. When up to 3 mL of 1N HCl was added to the polymer solutions, there was no significant change in the pH, probably due to protonation of the primary amine groups. However, the titration curves significantly differed from one another after the addition of more than 3 mL of HCl. Beyond this point, the pH of PEI600-g-CHI and PEI1800-g-CHI decreased faster than the pH of either PEI1800 or PEI600. The pKa of PEI600-g-CHI, PEI1800-g-CHI, and CHI was 10.0, 10.1, and 6.5, respectively. However, PEI600-g-CHI and PEI1800-g-CHI showed better buffering capacity than CHI alone.
Solubility of PEI-g-CHI copolymer
CHI has been used as a non-viral gene delivery vector.34 Due to the low solubility of CHI-based non-viral vectors, which limits their transfection efficiency, we report the synthesis of PEI-g-CHI with an increased solubility under physiological conditions. To test the increase in water solubility, the transmittance of PEI-g-CHI at 600 nm was measured. The result showed that the water solubility of PEI-g-CHI was better than CHI (Figure 4).
To study the solubility of CHI and PEI-g-CHI, the transmittance of CHI and PEI-g-CHI at 600 nm was measured. PEI600-g-CHI and PEI1800-g-CHI showed an improvement in solubility as compared with nascent CHI. The pH at 50% transmittance (T) at 600 nm is denoted as pH50 and the pH where T is higher than 98% at the same wavelength is denoted as the cloud point pH.29 The cloud point pH values of CHI, PEI600-g-CHI, and PEI1800-g-CHI were, respectively, 6.59, 6.82, and 6.83 as listed in Table 1.
Characterization of PEI-g-CHI/DNA polyplexes
Gel retardation assay
The ability of polycations to interact with plasmid DNA is an important requirement for an effective gene delivery system.35 Therefore, we assessed the capability of PEI-g-CHI to condense with DNA using agarose gel electrophoresis. Free plasmid DNA migrated through the gel and resolved into various distinct bands corresponding to supercoiled and nicked circular forms of the plasmid. For the PEI-g-CHI copolymer, the migration of DNA was completely inhibited when the weight ratio of PEI-g-CHI to DNA was more than 1. Figure 5 illustrates the low fluorescence of PEI1800-g-CHI/DNA polyplexes at weight ratios larger than 5. PEI1800-g-CHI/DNA resulted in a more compact polyplex, which hindered the staining of DNA with EtBr.
Particle size and zeta potential
The particle size of polymer-DNA polyplexes is an important factor that influences the passage of polyplexes into the targeting sites. Polycation/DNA polyplexes primarily enter the cell by endocytosis and thus have a size requirement below 300 nm for maximum endocytosis by non-specialized cells.36,37 Measured by DLS, the average particle sizes of PEI600-g-CHI/DNA at weight ratios (polymer:DNA) of 1, 5, 10, 20, and 40 were 380±58, 281±56, 246±87, 249±36, and 208±16 nm, respectively (Figure 6A). At weight ratios of 10 and 20, PEI600-g-CHI/DNA polyplexes were small enough to facilitate endocytosis. The average particle sizes of PEI1800-g-CHI/DNA (at weight ratios of 1, 5, 10, 20, and 40) were 353±19, 195±17, 179±23, 173±22, and 185±13 nm, respectively. At the same weight ratios, the particle sizes of PEI1800-g-CHI/DNA were smaller than PEI600-g-CHI/DNA. Higher molecular weight PEI resulted in a more compact polyplex.
The zeta potentials of polyplexes at various weight ratios are shown in Figure 6B. At a weight ratio of 1:1, the zeta potentials of PEI600-g-CHI/DNA and PEI1800-g-CHI polyplexes were 5.9±1.9 mV and 21.6±0.3 mV, respectively. With increasing weight ratio, the zeta potentials of the polyplexes rapidly increased to positive values. A positive surface charge is necessary for binding of polyplexes to anionic cell surfaces, which facilitates cell uptake.38 TEM and size distribution images of PEI600-g-CHI/DNA and PEI1800-g-CHI/DNA at a weight ratio of 10:1 are shown in Figures 6C and 6D, respectively.
Cytotoxicity of PEI-g-CHI copolymer
The cytotoxicity of the synthesized PEI-g-CHI conjugate was evaluated by MTT assay using a 293T cell line. At a concentration below 250 μg/mL, the cell viabilities of the three polymers (PEI600-g-CHI, PEI1800-g-CHI, and CHI) were approximately 100% (Figure 7). The results indicated that all three kinds of polymers were not harmful to the 293T cell line at a concentration below 250 μg/mL. At higher concentrations, PEI600-g-CHI and PEI1800-g-CHI had less cytotoxicity than CHI. Because CHI must be dissolved in an acid solution, the toxicity is primarily from the acidity, which is harmful to the cells. Polymers with more positive charge are more toxic to cells due to interference with mitochondrial function.39 These results demonstrated that PEI600-g-CHI and PEI1800-g-CHI were more biocompatible than PEI-25K.
Cell transfection of PEI-g-CHI copolymer
GFP expression
To investigate transfection efficiency of the PEI-g-CHI copolymer, we performed GFP in vitro, as shown in Figure 8. Fluorescence-activated cell sorting analysis was used to determine the transfection rate. When the polymer:DNA weight ratio was 10:1, PEI600-g-CHI/DNA exhibited the highest transfection efficiency, ie, 33% of GFP-positive cells were transfected with PEI600-g-CHI at this weight ratio.
The average relative fluorescence units of PEI600-g-CHI/DNA at polymer:DNA weight ratios of 1, 5, 10, 20, and 40, were 901±135, 20,475±2,884, 41,485±9,173, 12,169±5,148, and 18,610±3,270, respectively. The average relative fluorescence units of PEI1800-g-CHI/DNA, at weight ratios of 1, 5, 10, 20, and 40, were 1,946±359, 11,721±1,489, 6,235±762, 5,330±366, and 5,083±285, respectively. The maximal GFP expression of PEI600-g-CHI/DNA was at the weight ratio of 10, which was 3.5-fold higher than the maximal GFP expression of PEI1800-g-CHI/DNA at the weight ratio of 5. These results were in accordance with the GFP expression observed using fluorescence microscopy.
Water solubility, particle size, and surface charge of the polyplexes are key elements for successful gene transfection. The CHI of pKa is 6.5, which limits its solubility in acidic solutions and leads to poor gene transfection efficiency at physiological conditions.15 The water solubility of PEI600-g-CHI and PEI1800-g-CHI polymers are better than that of CHI. The particle sizes of PEI600-g-CHI/DNA and PEI1800-g-CHI/DNA polyplexes at weight ratios of 5–40 were within the scope of 173–281 nm, which allowed it to be easily taken up by the cell.36 The zeta potential of PEI600-g-CHI/DNA and PEI1800-g-CHI/DNA polyplexes at weight ratios of 5–40 were 6–39 mV with increasing weight ratio. A positive surface charge is necessary for binding of polyplexes to anionic cell surfaces, which facilitates cell uptake.38
Intracellular uptake of PEI600-g-CHI/DNA polyplexes
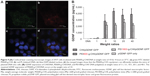
To visualize the intracellular uptake of the PEI600-g-CHI/DNA polyplexes, PI-labeled plasmid DNA, FITC-labeled PEI600-g-CHI, and DAPI were used. Figure 9A shows the uptake of the PEI600-g-CHI/pDNA polyplexes after 4 hours of transfection. Green dots of FITC-labeled PEI600-g-CHI were seen mainly in the cytoplasm, together with the red dots of PI-labeled plasmid DNA. Synchronized green dots with red dots were seen as yellow dots on the merged images, indicating that PEI600-g-CHI copolymers could efficiently bind with DNA. Meanwhile, a few green dots and red dots were present in the cytoplasm, indicating that DNA can be efficiently released from the polyplexes. These images revealed that the PEI600-g-CHI copolymers could efficiently mediate entry of plasmid DNA into cells.
GDNF expression
The GDNF expression of the PEI-g-CHI copolymer in vitro is shown in Figure 9B. The average GDNF concentrations of PEI600-g-CHI/DNA, at polymer:DNA weight ratios of 1, 5, 10, 20, and 40, were 13±5, 2,802±1,007, 4,346±448, 3,741±945, and 3,864±880 pg/mL, respectively. The average relative fluorescence units of PEI1800-g-CHI/DNA, at weight ratios of 1, 5, 10, 20, and 40, were 112±19, 2,140±403, 2,170±279, 2,560±506, and 2,165±278 pg/mL, respectively. The maximal GDNF expression of PEI600-g-CHI/DNA was at the weight ratio of 10, which was 1.7-fold higher than the maximal GDNF expression of PEI1800-g-CHI/DNA.
Conclusion
In this study, PEI600-g-CHI and PEI1800-g-CHI copolymer gene carriers were successfully synthesized and investigated for their physicochemical properties, biocompatibility, and gene transfection efficiency. The conjugation of PEI to CHI not only yielded a water-soluble polymer, but also increased CHI biocompatibility. PEI1800-g-CHI/DNA resulted in a more compact polyplex, which decreased DNA release. Therefore, PEI600-g-CHI proved superior to both CHI and PEI1800-g-CHI for gene transfection. In vitro GDNF gene delivery to 293T cells using PEI600-g-CHI resulted in significantly higher GDNF expression than GDNF gene delivery using either PEI1800-g-CHI or CHI. The low toxicity and high transfection efficiency of PEI600-g-CHI make it ideal for application to GDNF gene therapy, which is necessary for the potential treatment of Parkinson’s disease.
Acknowledgments
This work was supported by the Veterans General Hospital (grant no. VGHUST101-G6-1-1) and Chang Gung Memorial Hospital (grant no. CMRPG3C0101). The authors would like to thank Microscope Core Laboratory of Linkou Chang Gung Memorial Hospital and Ms. Wu Chia-Jung for their aide in confocal imaging.
Disclosure
The authors report no conflicts of interest in this work.
References
Hu FQ, Chen WW, Zhao MD, Yuan H, Du YZ. Effective antitumor gene therapy delivered by polyethylenimine-conjugated stearic acid-g-chitosan oligosaccharide micelles. Gene Ther. 2013;20(6):597–606. | ||
Huh MS, Lee SY, Park S, et al. Tumor-homing glycol chitosan/polyethylenimine nanoparticles for the systemic delivery of siRNA in tumor-bearing mice. J Control Release. 2010;144(2):134–143. | ||
Oliveira H, Pires LR, Fernandez R, Martins MC, Simões S, Pêgo AP. Chitosan-based gene delivery vectors targeted to the peripheral nervous system. J Biomed Mater Res A. 2010;95(3):801–810. | ||
Muramatsu S. Gene therapy for Parkinson disease. Nihon Rinsho. 2010;68 Suppl 8:646–649. Japanese. | ||
Park TG, Jeong JH, Kim SW. Current status of polymeric gene delivery systems. Adv Drug Deliv Rev. 2006;58(4):467–486. | ||
Wu TL, Ertl HC. Immune barriers to successful gene therapy. Trends Mol Med. 2009;15(1):32–39. | ||
Marshall E. Gene therapy death prompts review of adenovirus vector. Science. 1999;286(5448):2244–2245. | ||
Kato T, Iwamoto K, Ando H, et al. Synthetic cationic amphiphile for liposome-mediated DNA transfection with less cytotoxicity. Biol Pharm Bull. 1996;19(6):860–863. | ||
Anderson WF. Human gene therapy. Nature. 1998;392(6679 Suppl): 25–30. | ||
Köping-Höggård M, Tubulekas I, Guan H, et al. Chitosan as a nonviral gene delivery system. Structure-property relationships and characteristics compared with polyethylenimine in vitro and after lung administration in vivo. Gene Ther. 2001;8(14):1108–1121. | ||
Jiang HL, Kim YK, Arote R, et al. Chitosan-graft-polyethylenimine as a gene carrier. J Control Release. 2007;117(2):273–280. | ||
Toh EK, Chen HY, Lo YL, Huang SJ, Wang LF. Succinated chitosan as a gene carrier for improved chitosan solubility and gene transfection. Nanomedicine. 2011;7(2):174–183. | ||
Kadiyala I, Loo Y, Roy K, Rice J, Leong KW. Transport of chitosan-DNA nanoparticles in human intestinal M-cell model versus normal intestinal enterocytes. Eur J Pharm Sci. 2010;39(1–3):103–109. | ||
Dai H, Jiang X, Tan GC, et al. Chitosan-DNA nanoparticles delivered by intrabiliary infusion enhance liver-targeted gene delivery. Int J Nanomedicine. 2006;1(4):507–522. | ||
Bowman K, Leong KW. Chitosan nanoparticles for oral drug and gene delivery. Int J Nanomedicine. 2006;1(2):117–128. | ||
Lo YL, Sung KH, Chiu CC, Wang LF. Chemically conjugating polyethylenimine with chondroitin sulfate to promote CD44-mediated endocytosis for gene delivery. Mol Pharm. 2013;10(2):664–676. | ||
Neu M, Fischer D, Kissel T. Recent advances in rational gene transfer vector design based on poly(ethylene imine) and its derivatives. J Gene Med. 2005;7(8):992–1009. | ||
Zhang L, Chen Z, Li Y. Dual-degradable disulfide-containing PEI-Pluronic/DNA polyplexes: transfection efficiency and balancing protection and DNA release. Int J Nanomedicine. 2013;8:3689–3701. | ||
Boussif O, Lezoualc’h F, Zanta MA, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92(16):7297–7301. | ||
Shi S, Guo Q, Kan B, et al. A novel poly(epsilon-caprolactone)-pluronic-poly(epsilon-caprolactone) grafted polyethyleneimine(PCFC-g-PEI), Part 1, synthesis, cytotoxicity, and in vitro transfection study. BMC Biotechnol. 2009;9:65–76. | ||
Fernandes JC, Qiu X, Winnik FM, et al. Linear polyethylenimine produced by partial acid hydrolysis of poly(2-ethyl-2-oxazoline) for DNA and siRNA delivery in vitro. Int J Nanomedicine. 2013;8:4091–4102. | ||
Shastry BS. Parkinson disease: etiology, pathogenesis and future of gene therapy. Neurosci Res. 2001;41(1):5–12. | ||
Sajadi A, Bensadoun JC, Schneider BL, Lo Bianco C, Aebischer P. Transient striatal delivery of GDNF via encapsulated cells leads to sustained behavioral improvement in a bilateral model of Parkinson disease. Neurobiol Dis. 2006;22(1):119–129. | ||
Kells AP, Eberling J, Su X, et al. Regeneration of the MPTP-lesioned dopaminergic system after convection-enhanced delivery of AAV2-GDNF. J Neurosci. 2010;30(28):9567–9577. | ||
Bolliet C, Bohn MC, Spector M. Non-viral delivery of the gene for glial cell line-derived neurotrophic factor to mesenchymal stem cells in vitro via a collagen scaffold. Tissue Eng Part C Methods. 2008;14(3): 207–219. | ||
Li ZT, Guo J, Zhang JS, et al. Chitosan-graft-polyethylenimine with improved properties as a potential gene vector. Carbohyd Polym. 2010;80(1):254–259. | ||
Kircheis R, Wightman L, Wagner E. Design and gene delivery activity of modified polyethylenimines. Adv Drug Deliver Rev. 2001;53(3):341–358. | ||
Kubota N, Tatsumoto N, Sano T, Toya K. A simple preparation of half N-acetylated chitosan highly soluble in water and aqueous organic solvents. Carbohydr Res. 2000;324(4):268–274. | ||
Mao S, Shuai X, Unger F, Simon M, Bi D, Kissel T. The depolymerization of chitosan: effects on physicochemical and biological properties. Int J Pharm. 2004;281(1–2):45–54. | ||
Lou YL, Peng YS, Chen BH, Wang LF, Leong KW. Poly(ethylene imine)-g-chitosan using EX-810 as a spacer for nonviral gene delivery vectors. J Biomed Mater Res A. 2009;88(4):1058–1068. | ||
Malhotra M, Tomaro-Duchesneau C, Prakash S. Synthesis of TAT peptide-tagged PEGylated chitosan nanoparticles for siRNA delivery targeting neurodegenerative diseases. Biomaterials. 2013;34(4): 1270–1280. | ||
Intra J, Salem AK. Characterization of the transgene expression generated by branched and linear polyethylenimine-plasmid DNA nanoparticles in vitro and after intraperitoneal injection in vivo. J Control Release. 2008;130(2):129–138. | ||
Huang SJ, Wang TP, Lue SI, Wang LF. Pentablock copolymers of pluronic F127 and modified poly(2-dimethyl amino)ethyl methacrylate for internalization mechanism and gene transfection studies. Int J Nanomedicine. 2013;8:2011–2027. | ||
Mansouri S, Lavigne P, Corsi K, Benderdour M, Beaumont E, Fernandes JC. Chitosan-DNA nanoparticles as non-viral vectors in gene therapy: strategies to improve transfection efficacy. Eur J Pharm Biopharm. 2004;57(1):1–8. | ||
Khan M, Ang CY, Wiradharma N, et al. Diaminododecane-based cationic bolaamphiphile as a non-viral gene delivery carrier. Biomaterials. 2012;33(18):4673–4680. | ||
Liu YM, Reineke TM. Hydroxyl stereochemistry and amine number within poly(glycoamidoamine)s affect intracellular DNA delivery. J Am Chem Soc. 2005;127(9):3004–3015. | ||
Wu HC, Wang TW, Bohn MC, Lin FH, Spector M. Novel magnetic hydroxyapatite nanoparticles as non-viral vectors for the glial cell line-derived neurotrophic factor gene. Adv Funct Mater. 2010;20(1): 67–77. | ||
Petersen H, Fechner PM, Martin AL, et al. Polyethylenimine-graft-poly(ethylene glycol) copolymers: influence of copolymer block structure on DNA complexation and biological activities as gene delivery system. Bioconjug Chem. 2002;13(4):845–854. | ||
Fischer D, Bieber T, Li Y, Elsasser HP, Kissel T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm Res. 1999;16(8):1273–1279. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.