Back to Journals » Cancer Management and Research » Volume 11
Gephyrin suppresses lung squamous cell carcinoma development by reducing mTOR pathway activation
Authors Zhang X, Cheng D, Liu Y, Wu Y, He Z
Received 6 February 2019
Accepted for publication 24 April 2019
Published 7 June 2019 Volume 2019:11 Pages 5333—5341
DOI https://doi.org/10.2147/CMAR.S204358
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Antonella D'Anneo
Xiang Zhang,1,2 Dezhi Cheng,1,2 Yu Liu,1,2 Yuanbo Wu,2 Zhifeng He1,2
1Department of Thoracic Surgery, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, 325000, People’s Republic of China; 2Department of Cardiothoracic Surgery, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, 325000, People’s Republic of China
Background: The mTOR pathway is altered in a multitude of cancers, including lung cancer; however, abnormal activation in this pathway is less common in lung adenocarcinoma (LUAD) than in lung squamous cell carcinoma (LUSC). Gephyrin is a highly conserved and widely expressed ancient protein in vertebrate tissues. Its role and molecular mechanism in lung cancer development are largely unknown.
Method: We analyzed the expression profile of gephyrin and overall survival rates in LUAD and LUSC. The LUSC cells (H520 and SK-MES-1) were transfected with pLV-gephyrin to establish gephyrin stable overexpression cell lines. Real-time quantitative PCR and Western blot were performed to detect the mRNA and protein levels. The cell growth and cell cycle were detected by the MTT assay and flow cytometry. Finally, a xenograft tumor model was established to determine cell tumorigenesis in vivo.
Results: Our results show that gephyrin was reduced in LUAD and LUSC, and its low expression in LUSC patients indicated poor prognosis. Gephyrin overexpression suppressed LUSC cell proliferation, arrested cell cycle progression, and decreased the expression of cell-cycle related proteins such as cyclin D1, cyclin-dependent kinase-2 (CDK2), and proliferation-related protein proliferating cell nuclear antigen (PCNA). Conversely, knockdown of gephyrin promoted LUSC cell growth. Moreover, gephyrin reduced mTOR pathway activation to inhibit cyclin D1 and CDK2 translation. Mechanistically, gephyrin suppressed mTOR pathway activation by promoting mTOR degradation. Furthermore, gephyrin overexpression suppressed LUSC tumorigenesis.
Conclusion: Gephyrin suppressed LUSC development by reducing mTOR pathway activation, implicating gephyrin as a potential molecular target for LUSC management.
Keywords: gephyrin, lung squamous cell carcinoma, mTOR pathway, degradation
Introduction
Lung cancer is the second and the first most commonly diagnosed cancer in the USA and the People's Republic of China, respectively.1,2 It has the highest mortality rate of all cancers in both the USA and the People's Republic of China.1,2 In 2019, an estimated 228,150 people will be newly diagnosed and an estimated 142,670 people will die of lung cancer in the USA.1 Approximately 85% of lung cancers are non-small-cell lung cancer (NSCLC) and the others are small-cell lung cancer (SCLC).3 Lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) are the major two types of NSCLC. A large amount of evidence shows that tobacco smoking and exposure to asbestos, radon, and other potential carcinogens are key risk factors for lung cancer.4 Although some progress has been made in the study of lung cancer in the past few decades, the definite molecular mechanisms of lung cancer development remain largely unclear.
In the past few decades, the contributions of Ras, phosphoinositide 3-kinase (PI3K), and mTOR abnormal activation to lung tumor growth, maintenance, and chemoresistance have been extensively studied.5 A great deal of evidence also suggests that receptor tyrosine kinases (RTKs) and their downstream molecules are dysfunctional in lung cancer.6 In the case of NSCLC, targeted compounds which inhibit epidermal growth factor receptor (EGFR) have successfully reduced cancer development. However, the number of patients benefiting from targeted therapy is still not as huge as initially expected.4 More effective methods urgently need to be developed for lung cancer treatment.
Gephyrin (GPHN; from the Greek word for “bridge”) is a highly conserved and widely expressed neuronal receptor assembly protein which links with membrane-associated receptor molecules with cytoskeletal microfilaments in vertebrate tissues.7–10 Deletion of the GPHN gene in mice revealed that gephyrin regulates molybdenum cofactor biosynthesis, implying that the main functions of gephyrin in neurons are linked to the control of inhibitory neurotransmission.11,12 Eguchi et al reported that gephyrin can convert mixed-lineage leukemia (MLL) to an oncogene by fusion MLL-GPHN.13 However, the function of gephyrin in other cancers, such as lung cancer, is largely unknown.
Here, we report that gephyrin expression was lower in human NSCLC specimens than in the surrounding non-tumorous tissues, and gephyrin suppressed LUSC development via reduction of the mTOR pathway.
Materials and methods
Cell culture
LUSC cell lines H520 and SK-MES-1, and HEK293T cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). H520 cells were cultured in F12, SK-MES-1 cells were cultured in MEM, and HEK29T cells were cultured in DMEM supplemented with 10% FBS (HyClone), and 100 U/mL penicillin and 100 mg/mL streptomycin, and were maintained in a humidified incubator with 5% CO2 at 37°C.
Reagents and antibodies
Cycloheximide (CHX) and MG132 were obtained from Sigma Aldrich (St Louis, MO, USA) and dissolved in ethanol and DMSO, respectively. Antibodies for gephyrin, cyclin-dependent kinase 2 (CDK2), cyclin D1, proliferating cell nuclear antigen (PCNA), mTOR, 4E binding protein-1 (4E-BP1), p-4E-BP1 (Ser65), eukaryotic initiation factor 4E (eIF4E), p-eIF4E (Ser209), ribosomal S6 kinase (S6K), p-p70S6K (Thr389), ribosomal S6 (S6), and p-S6 (Ser235/236) were purchased from Cell Signaling Technologies. β-Actin antibody was purchased from Sigma.
Bioinformatic analysis of clinical data
The lung cancer data set was obtained from The Cancer Genome Atlas (TCGA) database. The expression of gephyrin was assessed by Gene Expression Profiling Interactive Analysis (GEPIA) (
Constructs and stable cell lines
Gephyrin cDNA was cloned into the pLV vector and pCMV-myc vector, and mTOR cDNA was cloned into the pCNDA3-HA vector. Constructs were transfected with lipofectamine2000 following the manufacturer’s instructions. The lentivirus was packaged following the standard protocol. To establish stable gephyrin overexpression cell lines, H520 and SK-MES-1 cells were transfected with pLV-gephyrin and pLV control vector lentivirus, respectively. Stably transfected cells were selected with 1 mg/mL puromycin, and the mono-clone was selected.
MTT assay
Gephyrin stable overexpression or control H520 and SK-MES-1 cells were seeded in 96-well plates at a density of 2,000 per well in 100 μL of the corresponding medium. The cells were grown for different times (0, 24, 48, 72, and 96 hours). Subsequently, they were treated with a fresh solution of MTT (5 mg/mL) for 4 hours at 37°C. The purple formazan crystals were ultimately solubilized with DMSO solution, and absorbance was recorded using a multiwell plate reader at 490 nm.
Cell cycle analysis
Gephyrin overexpressed and control H520 and SK-MES-1 cells were harvested, washed with PBS, then fixed in ice-cold 70% (v/v) ethanol overnight at 4°C. Before analysis, cells were washed with PBS and stained with propidium iodide according to the manufacturer’s instructions. The samples were analyzed by flow cytometry (FACSCalibur flow cytometer; BD Biosciences, San Jose, CA, USA) using Cell Quest software.
Western blot analysis
Cells or tumor tissues were homogenized in RIPA buffer, and debris was removed by centrifugation at 12,000 rpm for 10 minutes at 4°C. The protein concentrations in all samples were determined by the Bradford protein assay kit (Bio-Rad, Hercules, CA, USA). After addition of the sample loading buffer, protein samples were electrophoresed and then transferred to poly-vinylidene difluoride transfer membranes. The blots were blocked for 1 hour at room temperature with fresh 5% non-fat milk in Tris-buffered saline with Tween (TBST) and then incubated with the specific primary antibody in TBST overnight at 4°C. Following three washes with TBST, the blots were incubated with the appropriate horseradish peroxidase-conjugated secondary antibody for 1 hour, and the immunoreactive bands were visualized using the ECL kit (Bio-Rad, Hercules, CA, USA).
RNA extraction and real-time quantitative PCR
Cells were homogenized in TRIzol reagent (Invitrogen, New York, NY, USA) and the total RNA was isolated according to the manufacturer’s instructions. Then, 2 μg total RNA was reversed to mRNA for qPCR according to the manufacturer’s instructions (RT-PCR system; Toyobo, Osaka, Japan). qRT-PCR was performed using the SYBR Green (Bio-Rad) according to the manufacturer’s instructions, with the following primers for GPHN: sense primer CCGTGTCGGAGTCCTTACAG; anti-sense primer AGTCCCACCCAACAAAGAAGG; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH): sense primer TGTGGGCATCAATGGATTTGG and anti-sense primer ACACCATGTATTCCGGGTCAAT (Invitrogen). GAPDH was used as the inner control. The relative mRNA levels were quantified using the 2−ΔΔCt method.
siRNA knockdown
The special gephyrin siRNA (IDs 136977 and 136978) was purchased from Invitrogen (Carlsbad, CA, USA). Negative universal control (Invitrogen) was used as the control. Cells were seeded at 5×105/well into six-well plates for overnight incubation, and transfected with gephyrin or control siRNAs (80 nM) using lipofectamine RNAiMAX (Invitrogen). Forty-eight hours after transfection, cells were harvested, and the degree of knockdown was detected by Western blot analysis of gephyrin proteins.
Tumor xenograft experiments
The xenograft tumor model protocol was approved by the Institutional Animal Care and Use Committee of the Laboratory Animal Center of Wenzhou Medical University. Male nude mice, 4–6 weeks old, were purchased from the Laboratory Animal Center of Wenzhou Medical University. All mice were housed with a 12:12-hour light-dark cycle at room temperature, and fed with a standard rodent diet and water. The animals were acclimatized to the laboratory for at least 1 week before use in experiments. All animal care and welfare was performed in accordance with “The Detailed Rules and Regulations of Medical Animal Experiments Administration and Implementation” (document no. 1998-55; Ministry of Public Health, People's Republic of China). Nude mice were subcutaneously injected in both flanks with 2×106 H520 overexpression cells or control cells. Ten days after injection, the volume of the tumors was monitored and calculated following the formula: Volume = Length × Width2× 0.52. Tumors were harvested and weighed, and then the tumors were homogenized in RIPA for Western blot analysis.
Statistical analysis
All data are expressed as mean ± SEM, obtained from more than three independent experiments, and analyzed by GraphPad Prism 5.0 (GraphPad Software, San Diego, CA, USA). Statistically significant differences (p<0.05) were examined using the Student's t-test and two-way ANOVA.
Results
Gephyrin is reduced in lung cancer and indicates poor prognosis in LUSC patients
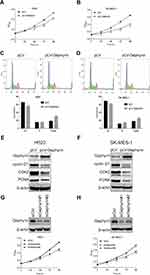
To reveal the expression profile of gephyrin in human lung cancer, we examined the expression of gephyrin in human lung cancer and the surrounding non-tumorous lung tissues. The data from TCGA showed that the mRNA levels of gephyrin in LUAD and LUSC were remarkably lower than those in normal lung tissues (Figure 1A). The overall survival rate was not significantly different between patients who expressed low or high levels of gephyrin in LUAD (Figure 1B); however, the expression of low levels of gephyrin indicated poor prognosis in LUSC (Figure 1C). These results indicate that the reduction of gephyrin may promote LUSC progression.
Gephyrin suppresses LUSC cell proliferation
To determine the role of gephyrin in LUSC cell proliferation, the gephyrin cDNA was cloned into the pLV vector to establish stable overexpression of gephyrin human LUSC cell lines H520 and SK-MES-1. First, the MTT assay was used to generate the lung cancer cell growth curve. As shown in Figure 2A and B, overexpression of gephyrin reduced the growth rates of H520 (Figure 2A) and SK-MES-1 (Figure 2B) cells. It has been reported that the cell cycle is one of the regulators which control cell growth.14 Then, we detected whether gephyrin inhibited cell growth through controlling the cell cycle. Cell flow cytometry was used to measure the effects of gephyrin on the cell cycle. Overexpression of gephyrin arrested the cell cycle at G1 phase in H520 (Figure 2C) and SK-MES-1 (Figure 2D) cells. Furthermore, we also detected the protein levels of cell-cycle regulated genes, such as cyclin D1 and CDK2. The Western blot results showed that overexpression of gephyrin inhibited the expression of cyclin D1 and CDK2 in H520 (Figure 2E) and SK-MES-1 (Figure 2F) cells. The proliferation marker PCNA was also suppressed by gephyrin in H520 (Figure 2E) and SK-MES-1 (Figure 2F) cells. In addition, knockdown of gephyrin by siRNA promoted H520 (Figure 2G) and SK-MES-1 (Figure 2H) cell growth. These data demonstrate that gephyrin can inhibit LUSC cell growth through regulating the cell cycle.
Overexpression of gephyrin inhibits the mTOR pathway
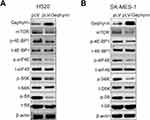
It has been reported that mTOR can regulate cell cycle progression through its cell growth effectors S6K and 4E-BP1/eIF-4E.14 The mTOR pathway can regulate the translation of cyclin D1 and CDK2. So, we proposed that gephyrin controls the cell cycle through the mTOR pathway. We used the gephyrin stable overexpression LUSC cell lines to detect the effects of gephyrin on the mTOR pathway. As shown in Figure 3, overexpression of gephyrin could suppress the phosphorylation of 4E-BP1, eIF4E, S6K, and SK, as well as reduce the expression of mTOR in H520 (Figure 3A) and SK-MES-1 (Figure 3B) cells. These results indicate that gephyrin suppressed the mTOR pathway to control the cell cycle.
Gephyrin promotes mTOR degradation
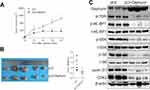
To identify the mechanism of gephyrin in reducing mTOR expression, we first detected the mRNA level of mTOR in H520 and SK-MES-1 cells. Overexpression of gephyrin did not change the mRNA levels of mTOR (Figure 4A). This means that gephyrin may regulate mTOR expression through post-translational levels. Then, we detected the role of gephyrin in mTOR stability. We cotransfected HA-mTOR and myc-gephyrin plasmids into HEK293T cells for 48 hours, then treated them with CHX for different times. The protein levels of mTOR were measured by Western blot. Gephyrin significantly reduced the stability of mTOR (half-life [t1/2]=1.9 hours in the gephyrin overexpression group and t1/2=11.2 hours in the control group) (Figure 4B). Lastly, we detected whether gephyrin promoted mTOR degradation through enhancing mTOR ubiquitin (Ub). HA-Ub, HA-mTOR, and myc-gephyrin plasmids were cotransfected into HEK293T cells for 48 hours, then treated with MG132 for 8 hours. The ubiquitin of mTOR was measured by Western blot. As shown in Figure 4C, gephyrin enhanced mTOR ubiquitin. These results suggest that gephyrin promoted mTOR degradation by enhancing its ubiquitin.
Overexpression of gephyrin suppresses H520 cell tumorigenesis
The data presented in the previous subsections demonstrate that gephyrin suppressed LUSC cell growth through inhibiting the mTOR pathway. Then, a xenograft tumor model was used to detect the effects of gephyrin overexpression on LUSC cell growth in vivo. Gephyrin overexpression and control H520 cells were subcutaneously injected into nude mice. Tumor sizes were monitored every 3 days from day 10 after cell injection, and tumors were harvested at the experimental endpoint. As shown in Figure 5A and B, the tumors in the gephyrin overexpression group grew much more slowly and weighed much less compared with those in the control group. Furthermore, gephyrin overexpression also suppressed mTOR expression, inhibited the phosphorylation of 4E-BP, S6K, and S6, and reduced the expression of cell cycle protein CDK2 and cyclin D1 in H520 tumors (Figure 5C). Collectively, these data indicate that gephyrin suppressed LUSC cell growth both in vitro and in vivo, at least in part through down-regulating the mTOR pathway.
Discussion
In the present study, we clearly demonstrate the key role of gephyrin in LUSC development (Figure 6): 1) a marked down-regulation of gephyrin in LUSC was identified by analyzing TCGA data set; 2) overexpression of gephyrin significantly reduced LUSC cell growth by arresting the cell cycle at G1 phase; 3) overexpression of gephyrin inhibited the mTOR pathway through controlling the translation of cell-cycle related genes, such as cyclin D1 and CDK2; 4) overexpression of gephyrin promoted mTOR degradation; and 5) overexpression of gephyrin suppressed LUSC tumorigenesis.
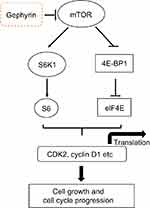 | Figure 6 Schematic illustration of the underlying mechanism of gephyrin reducing LUSC development.Abbreviations: CDK2, cyclin-dependent kinase 2; LUSC, lung squamous cell adenocarcinoma. |
Gephyrin, a highly conserved and widely expressed protein, plays an essential role in regulating neuronal function, including molybdenum cofactor synthesis,7,8,15,16 postsynaptic signaling transduction,17–20 GABAergic synapse formation,21,22 and GABAergic synaptic plasticity.21,23,24 GPHN mutations may cause some neurological diseases (eg, molybdenum cofactor deficiency,25 stiff-person syndrome,26 and hyperekplexia27). The functions of gephyrin in other diseases are largely unknown. Occasional reports have indicated that gephyrin can convert MLL to an oncogene by fusion MLL-GPHN.13 We proposed that gephyrin may have a function in the development of other cancers, such as lung cancer. As shown by the data in this paper, we found that gephyrin was reduced in LUSC, and suppressed LUSC cell growth in vitro (Figure 2) and in vivo (Figure 5). Gephyrin significantly suppressed cancer cell proliferation, raising the question of whether gephyrin influences cell proliferation physiologically. Our data show that gephyrin did not influence the cell proliferation of normal lung cell Base-2B (data no shown). Therefore, gephyrin plays a key role in suppressing LUSC development; however, it has no effects on normal cell proliferation.
The target of rapamycin (TOR) was originally identified by mutations which confer resistance to the growth-inhibitory properties of rapamycin.28 Cell growth is appropriately controlled in both time and space. When nutrients and other growth stimuli are present, TOR is active and the cell maintains a vigorous rate of ribosome biogenesis, translation initiation, and nutrient import to enhance macromolecular synthesis and thereby increase in size and mass.29 The mTOR pathway is altered in many human cancers.5,30,31 mTOR has been identified as a downstream target of the PI3K/AKT pathway, and mediates the effects of cell proliferation and survival in lung cancer.4,6 Targeted inhibition of the mTOR pathway can prevent tumor progression. In this study, we observed that overexpression of gephyrin significantly reduced the expression of cyclin D1 and CDK2, indicating that gephyrin suppressed LUSC development, at least in part, by negatively mediating the expression of these proteins. Because cyclin D1 and CDK2 are the target genes of the mTOR pathway, which is frequently abnormally activated in LUSC, we proposed that gephyrin could suppress LUSC development by decreasing mTOR activation.14 The present results demonstrate that gephyrin suppresses the mTOR pathway by reducing the protein levels of mTOR to inhibit cell-cycle related protein translation (Figure 3). Ubiquitin is one of major factors in protein degradation and plays a central role in the cell life cycle.32 Therefore, we hypothesized that gephyrin may reduce mTOR protein levels by promoting mTOR ubiquitylation, and our results supported this hypothesis (Figure 4C). Therefore, developing gephyrin agonists to block the mTOR pathway could be an attractive therapeutic approach for treating LUSC.
Conclusion
In summary, our study demonstrated that gephyrin reduced LUSC development through reduction of the mTOR pathway, and overexpression of gephyrin could reduce cancer cell development. Therefore, gephyrin is a novel tumor suppressor in LUSC and a potent molecular target in LUSC treatment.
Abbreviation list
CDK2, cyclin-dependent kinase 2; CHX, cycloheximide; 4E-BP1, 4E binding protein-1, eIF4E, eukaryotic initiation factor 4E; GPHN, gephyrin; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MLL, mixed-lineage leukemia; NSCLC, non-small-cell lung cancer; PCNA, proliferating cell nuclear antigen; PI3K, phosphoinositide 3-kinase; RTK, receptor tyrosine kinase; S6, ribosomal S6; S6K, ribosomal S6 kinase; TCGA, The Cancer Genome Atlas; Ub, ubiquitin.
Acknowledgments
This work was supported by grants from the Natural Science Foundation of Zhejiang Province (LQ18H010004) and Wenzhou Public Welfare Science and Technology Plan (Y20160047).
Author contributions
XZ and ZH conceived and coordinated the study. XZ and ZH wrote the paper. XZ, DC, YL, YW, and ZH designed, performed, and analyzed the experiments. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians. 2019;69(1):7–34.
2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA. 2016;66(2):115–132.
3. Jackman DM, Johnson BE. Small-cell lung cancer. Lancet. 2005;366(9494):1385–1396. doi:10.1016/S0140-6736(05)67528-9
4. Marinov M, Fischer B, Arcaro A. Targeting mTOR signaling in lung cancer. Crit Rev Oncol Hematol. 2007;63(2):172–182. doi:10.1016/j.critrevonc.2007.04.002
5. Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441(7092):424–430. doi:10.1038/nature04869
6. Pisick E, Jagadeesh S, Salgia R. Receptor tyrosine kinases and inhibitors in lung cancer. Sci World J. 2004;4:589–604. doi:10.1100/tsw.2004.117
7. Nawrotzki R, Islinger M, Vogel I, Volkl A, Kirsch J. Expression and subcellular distribution of gephyrin in non-neuronal tissues and cells. Histochem Cell Biol. 2012;137(4):471–482. doi:10.1007/s00418-012-0914-7
8. Ogino K, Ramsden SL, Keib N, Schwarz G, Harvey RJ, Hirata H. Duplicated gephyrin genes showing distinct tissue distribution and alternative splicing patterns mediate molybdenum cofactor biosynthesis, glycine receptor clustering, and escape behavior in zebrafish. J Biol Chem. 2011;286(1):806–817. doi:10.1074/jbc.M110.125500
9. Kneussel M, Betz H. Receptors, gephyrin and gephyrin-associated proteins: novel insights into the assembly of inhibitory postsynaptic membrane specializations. J Physiol. 2000;525(Pt 1):1–9.
10. Prior P, Schmitt B, Grenningloh G, et al. Primary structure and alternative splice variants of gephyrin, a putative glycine receptor-tubulin linker protein. Neuron. 1992;8(6):1161–1170. doi:10.1016/0896-6273(92)90136-2
11. Schwarz G, Mendel RR, Ribbe MW. Molybdenum cofactors, enzymes and pathways. Nature. 2009;460(7257):839–847. doi:10.1038/nature08302
12. Smolinsky B, Eichler SA, Buchmeier S, Meier JC, Schwarz G. Splice-specific functions of gephyrin in molybdenum cofactor biosynthesis. J Biol Chem. 2008;283(25):17370–17379. doi:10.1074/jbc.M800985200
13. Eguchi M, Eguchi-Ishimae M, Greaves M. The small oligomerization domain of gephyrin converts MLL to an oncogene. Blood. 2004;103(10):3876–3882. doi:10.1182/blood-2003-11-3817
14. Fingar DC, Richardson CJ, Tee AR, Cheatham L, Tsou C, Blenis J. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol Cell Biol. 2003;24(1):200–216. doi:10.1128/MCB.24.1.200-216.2004
15. Reiss J, Lenz U, Aquaviva-Bourdain C, Joriot-Chekaf S, Mention-Mulliez K, Holder-Espinasse M. A GPHN point mutation leading to molybdenum cofactor deficiency. Clin Genet. 2011;80(6):598–599. doi:10.1111/j.1399-0004.2011.01709.x
16. Stallmeyer B, Schwarz G, Schulze J, et al. The neurotransmitter receptor-anchoring protein gephyrin reconstitutes molybdenum cofactor biosynthesis in bacteria, plants, and mammalian cells. Proc Natl Acad Sci USA. 1999;96(4):1333–1338. doi:10.1073/pnas.96.4.1333
17. Pfeiffer F, Graham D, Betz H. Purification by affinity chromatography of the glycine receptor of rat spinal cord. J Biol Chem. 1982;257(16):9389–9393.
18. Kirsch J, Langosch D, Prior P, Littauer UZ, Schmitt B, Betz H. The 93-kDa glycine receptor-associated protein binds to tubulin. J Biol Chem. 1991;266(33):22242–22245.
19. Barnard EA, Skolnick P, Olsen RW, et al. International union of pharmacology. XV. Subtypes of gamma-aminobutyric acidA receptors: classification on the basis of subunit structure and receptor function. Pharmacol Rev. 1998;50(2):291–313.
20. Durisic N, Godin AG, Wever CM, Heyes CD, Lakadamyali M, Dent JA. Stoichiometry of the human glycine receptor revealed by direct subunit counting. J Neurosci. 2012;32(37):12915–12920. doi:10.1523/JNEUROSCI.2681-12.2012
21. Tyagarajan SK, Ghosh H, Yevenes GE, et al. Regulation of GABAergic synapse formation and plasticity by GSK3beta-dependent phosphorylation of gephyrin. Proc Natl Acad Sci U S A. 2011;108(1):379–384. doi:10.1073/pnas.1011824108
22. Takahashi H, Katayama K, Sohya K, et al. Selective control of inhibitory synapse development by Slitrk3-PTPdelta trans-synaptic interaction. Nat Neurosci. 2012;15(3):
23. Tyagarajan SK, Ghosh H, Yevenes GE, et al. Extracellular signal-regulated kinase and glycogen synthase kinase 3beta regulate gephyrin postsynaptic aggregation and GABAergic synaptic function in a calpain-dependent mechanism. J Biol Chem. 2013;288(14):9634–9647. doi:10.1074/jbc.M112.442616
24. Luscher B, Fuchs T, Kilpatrick CL. GABAA receptor trafficking-mediated plasticity of inhibitory synapses. Neuron. 2011;70(3):385–409. doi:10.1016/j.neuron.2011.03.024
25. Reiss J, Gross-Hardt S, Christensen E, Schmidt P, Mendel RR, Schwarz G. A mutation in the gene for the neurotransmitter receptor-clustering protein gephyrin causes a novel form of molybdenum cofactor deficiency. Am J Hum Genet. 2001;68(1):208–213. doi:10.1086/316941
26. Butler MH, Hayashi A, Ohkoshi N, et al. Autoimmunity to gephyrin in Stiff-Man syndrome. Neuron. 2000;26(2):307–312. doi:10.1016/S0896-6273(00)81165-4
27. Rees MI, Harvey K, Ward H, et al. Isoform heterogeneity of the human gephyrin gene (GPHN), binding domains to the glycine receptor, and mutation analysis in hyperekplexia. J Biol Chem. 2003;278(27):24688–24696. doi:10.1074/jbc.M301070200
28. Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253(5022):905–909. doi:10.1126/science.1715094
29. Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. doi:10.1016/j.cell.2006.01.016
30. Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5(12):921–929. doi:10.1038/nrc1753
31. Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4(12):988–1004. doi:10.1038/nrd1876
32. Zheng N, Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem. 2017;86:129–157. doi:10.1146/annurev-biochem-060815-014922
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.