Back to Journals » Infection and Drug Resistance » Volume 11
Genotyping and antibiotic resistance properties of Helicobacter pylori strains isolated from human and animal gastric biopsies
Authors Ranjbar R , Chehelgerdi M
Received 17 September 2018
Accepted for publication 13 November 2018
Published 13 December 2018 Volume 2018:11 Pages 2545—2554
DOI https://doi.org/10.2147/IDR.S187885
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Reza Ranjbar,1 Mohammad Chehelgerdi2
1Molecular Biology Research Center, Systems Biology and Poisonings Institute, Baqiyatallah University of Medical Sciences, Tehran, Iran; 2Biotechnology Research Center, Shahr-e Kord Branch, Islamic Azad University, Shahr-e Kord, Iran
Purpose: The present study was done to assess the prevalence rate, antibiotic resistance pattern and genotyping status of the Helicobacter pylori strains isolated from human and animal gastric biopsy samples.
Patients and methods: A total of 1,150 gastric biopsy samples were randomly collected from humans (children and adults) and animals (cows, sheep and goats). All samples were subjected to culture, urease test and histopathologic examination. H. pylori isolates were also confirmed using the 16S rRNA gene PCR-amplification. Antibiotic resistance pattern was assessed by the disk diffusion method. Distribution of different genotypes was studied by PCR.
Results: The prevalence of H. pylori in gastric biopsy samples which were studied using urease test, culture and histological examination were 57.04%, 55.40% and 60.80%, respectively. Samples that were collected from adult humans (78%) and sheep (70%) had the highest prevalence of H. pylori strains, while those of goats (0.6%) and cows (4%) had the lowest. Findings of the culture method were confirmed using PCR-based amplification of 16S rRNA. Distribution of H. pylori among the gastric ulcers, duodenal ulcers, chronic gastritis gastric cancer and chronic cancer samples were 10.40%, 15.70%, 96.50%, 0.60% and 3.14%, respectively. H. pylori strains harbored the highest prevalence of resistance against ampicillin (74.4%), clarithromycin (63.4%), trimethoprim (61.5%) and metronidazole (61.5%). The most commonly detected genotypes among the H. pylori strains isolated from different types of biopsy samples were cagA (84.79%), vacA m2 (55.95%), vacA s1a (49.84%), cagE (48.58%), iceA1 (47.02%) and iceA2 (47.02%).
Conclusion: High prevalence of antibiotic resistance and virulent genotypes indicates an important public health issue. Similarities in antibiotic resistance and genotyping pattern of H. pylori strains isolated from humans and animals may show their similar routes of infection.
Keywords: Helicobacter pylori, virulence factor, antibiotic resistance, genotypes, clinical samples
Introduction
Helicobacter pylori (H. pylori) is a microaerophilic and Gram-negative spiral coccoid flagellated bacterium with 2–4 µm length and 0.5–1 µm width. It is known as one of the main causative agents of duodenal ulcer, peptic ulcer disease, gastric adenocarcinoma, type B gastritis and gastric B-cell lymphoma.1–3 Human stomach is considered as a main reservoir of H. pylori strains.1,2,4 In keeping with this, animals such as cows, sheep and goats may play an imperative role in the transmission of H. pylori infections to humans.5–7 The prevalence of H. pylori infections in some countries such as Iran is very high, and epidemiological studies showed that more than 50% of the human population are affected with H. pylori strains.8,9
H. pylori bacteria colonize the superficial parts of the gastric mucosa.8,10,11 It acts as an invasive bacterium and causes several gastrointestinal disorders and lesions in the gastric mucosa. Different pathogenic pathways have been developed for H. pylori infection. Signaling pathways were shown to be perturbed in gastric epithelial cells by virulence factors of H. pylori bacteria.12–14 Some of the most important virulence genes of this bacterium are the outer inflammatory protein (oip), vacuolating cytotoxin A (vacA), induced by contact with the epithelium antigen (iceA), blood group antigen-binding adhesion (babA) and cag PAI (cag pathogenicity island).15 Colonization, adhesion and invasion of H. pylori strains in gastric epithelial cells are facilitated by the presence of different virulence genes.16,17 vacA and cytotoxin associated gene A (cagA) are two important virulence genes with high importance in the pathogenicity of H. pylori infections.18,19 The vacA gene is polymorphic, comprising variable signal regions (type s1 or s2) and mid-regions (type m1 or m2). The s1 type is additionally divided into s1a, s1b and s1c and the m1 into m1a and m1b subtypes. The cagA gene has been detected in the severe cases of gastrointestinal disorders and peptic ulcers.16,17 The iceA gene has two main allelic variants: iceA1 and iceA2. A small percentage of the H. pylori genome is predicted to encode outer membrane proteins (OMPs). The oipA gene plays an important role in efficient colonization of H. pylori into the mucosa.20 Genotyping using these virulence markers is considered as one of the best approaches to study the correlations between H. pylori isolates from different samples.7
Antibiotic therapy is one of the best aspects of treatments for H. pylori infections. However, therapeutic options have become somewhat restricted because of the presence of severe resistance in some strains of this bacterium.6 Documented data disclosed that H. pylori strains isolated from clinical infections harbored high prevalence of resistance against different types of antibiotics including fluoroquinolone, tetracyclines, penicillins, aminoglycosides, sulfonamides and macrolides.7 Therefore, it is important to know the exact antibiotic resistance pattern of H. pylori strains isolated from human and animal clinical infections.
Data on the epidemiology and transmission of H. pylori is extremely significant in order to prevent its distribution and to identify high-risk populations, especially in areas that have high rates of infections such as Iran. Considering the indistinct epidemiological aspects of H. pylori in human and animal clinical samples and the high prevalence of H. pylori all-around the world, the present investigation was done in order to study the prevalence rate, genotyping patterns and antibiotic resistance pattern of H. pylori strains isolated from human and animal clinical samples.
Methods
Sample collection
Number of samples to be collected was obtained based on the prevalence data of H. pylori found in recent studies and using the following equation:
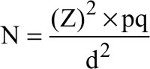
whereas p is the mean prevalence of H. pylori in recent studies, (Z)2 is the abscissa of the standard curve that cuts off an area α at the tails, d is the acceptable sampling error and N is the sample size.
The present cross-sectional study was conducted in the period between December 2016 and March 2017. Samples were collected from different parts of the Chaharmahal va Bakhtiari Province, Iran. Chaharmahal va Bakhtiari Province is located in an area of 16,411 km2 between 31 degrees and 9 minutes to 32 degrees and 48 minutes of north latitude and 49 degrees and 28 minutes to 51 degrees and 25 minutes of east longitude from Greenwich meridian (southwest of Iran).
Samples were randomly collected from the gastric biopsies of humans (children [≤14 years] [n=50] and adults [650 samples: 408 females and 242 males] and animals (450 samples: 150 cows, 150 sheep and 150 goats). All patients were referred to the Hajar Hospital for upper gastrointestinal endoscopy. Patients with a history of partial gastric resection or previous treatment for H. pylori infection were excluded. Gastric biopsy samples of antrum were taken from humans. Samples of animals were taken from slaughterhouses in the same region as the humans. Samples from the rumen of animals in size of 2×4×4 mm3 were obtained immediately after slaughtering. All samples were kept in transport medium consisting of thioglycolate with 1.3 g/L agar with 2% yeast extract and transmitted to the Biotechnology Research Center of the Islamic Azad University of Shahrekord Branch in cooler with ice packs.
Isolation of H. pylori
Three different methods were used for diagnosis of H. pylori strains in the studied samples. All samples were cultured on Brucella Agar (EMD Millipore, Billerica, MA, USA) containing 5 mg/L of campylobacter selective supplement (EMD Millipore), 30 mg/L of colistin methanesulfonate, 100 mg/L of cycloheximide, 30 mg/L of nalidixic acid, 30 mg/L of trimethoprim, 10 mg/L of vancomycin (Sigma-Aldrich Co., St Louis, MO, USA), amphotericin B, 7% sheep blood and 6% fetal calf serum (EMD Millipore). Media were then incubated at 37°C for 3–10 days on microaerophilic condition (85% N2, 10% CO2 and 5% O2). Grown bacteria were identified using colony morphology, urease activities and positive reactions to oxidase, catalase and Gram staining. For histology method, the specimens were fixed in 10% buffered formalin and then embedded in paraffin and 5 mm thick sections were cut. Serial sections (biopsies ranged from 2×4×4 mm3) were stained with hematoxylin–eosin and May–Giemsa for histologic examination. Using endoscopy assay, gastric cancer and peptic ulcer were identified, and also gastritis in the peptic ulcer or gastric malignancy were diagnosed by an expert professor.
DNA extraction and genotyping of H. pylori strains
Typical colonies were subcultured on Wilkins-Chalgren anaerobe broth (EMD Millipore) supplemented with and cultured in similar conditions mentioned above. All tubes were incubated for 7 days at 37°C with shaking under microaerophilic conditions. Then, genomic DNA was extracted from grown colonies using a DNA extraction kit for cells and tissues (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. After extraction, the DNA samples were quantified (NanoDrop, Thermo Fisher Scientific), their purity checked (A260/A280) and their concentrations adjusted to 50 ng/µL. The integrity of the DNA was evaluated on a 2% agarose gel stained with ethidium bromide (0.5 µg/mL) (Thermo Fisher Scientific). The DNA concentration was also estimated by spectrophotometric absorbance at 257 nm (DR5000, Hach Company, Loveland, CO, USA).21 The DNA was stored at –20°C pending subsequent PCR analysis.
Extracted DNA was amplified for the 16S rRNA gene (primers: HP-F: 5’-CTGGAGAGACTAAGCCCTCC-3’ and HP-R: 5’-ATTACTGACGCTGATTGTGC-3’).6 The genotypes of vacA (s1a, s1b, s1c, m1a, m1b and m2) and the presence of iceA1, iceA2, oipA, cagA, cagE and babA2 genotypes were determined using PCR technique. The primers used in this study are listed in Table 1. All PCR mixtures were prepared in a final volume of 25 µL containing 50 ng DNA from the samples served as the template for PCR performed in a thermal cycler (Mastercycler gradient, Eppendorf, Germany), 1 µM of each primer, 2 mM MgCl2, 5 µL of 10× PCR buffer, 200 µM dNTPs and 1 unit of Taq DNA polymerase (CinnaGen Co, Tehran, Iran). Thermal amplification of PCR products was performed at 95°C for 5 minutes and then for 32 cycles of 94°C for 1 minute, 61°C for 40 seconds, 72°C for 40 seconds and a final extension at 72°C for 5 minutes, with a final hold at 10°C in a PCR thermal cycler (Mastercycler gradient). The PCR amplified products (10 µL) were subjected to electrophoresis in 1.5% agarose gel in 1× TBE buffer at 80 V for 30 minutes stained with a solution of ethidium bromide (EMD Millipore). and examined under Ultra Violet illumination (Cleaver Scientific Ltd, Rugby, UK).
  | Table 1 PCR primers and PCR conditions used for genotypes of H. pylori strains in this study Notes: *Table adapted from Ranjbar R, Khamesipour F, Jonaidi-Jafari N, Rahimi E. Helicobacter pylori in bottled mineral water: genotyping and antimicrobial resistance properties. BMC Microbiol. 2016;16:1–10. © Ranjbar et al 2016.46 |
Antimicrobial resistance pattern
The pure cultures of H. pylori strains were employed for antibiotic susceptibility test. One strain from each H. pylori-positive sample was selected and then subjected to the Kirby-Bauer disc diffusion method using Mueller-Hinton agar (EMD Millipore) supplemented with 5% defibrinated sheep blood and 7% fetal calf serum, according to the Clinical Laboratory Standards Institute (CLSI 2015).22 The antimicrobial resistance pattern of H. pylori was measured against the widely used antibiotics in cases of H. pylori gastric ulcer. The following antimicrobial disks (Thermo Fisher Scientific) were used: metronidazole (5 µg), ampicillin (10 µg), erythromycin (5 µg), clarithromycin (2 µg), furazolidone (1 µg), levofloxacin (5 µg), amoxicillin (10 µg), trimethoprim (25 µg), moxifloxacin (5 µg), cefsulodin (30 µg), tinidazole (5 µg) and ciprofloxacin (5 µg) were used. After incubation at 37°C for 48 hours in a microaerophilic atmosphere (5% oxygen, 85% nitrogen and 10% CO2) using MART system (Anoxomat, Lichtenvoorde, the Netherlands), the susceptibility of the strains was measured against each antimicrobial agent. Results were construed in accordance with interpretive criteria provided by CLSI (2015). H. pylori ATCC 43504 was used as quality control organism in the antimicrobial susceptibility determination. The results were interpreted as resistant (R).
Statistical analyses
Data were transferred to Microsoft Excel spreadsheet (version 15; Microsoft Corporation, Redmond, WA, USA) for analysis. Using statistical software (version 16; SPSS Inc., Chicago, IL, USA), chi-squared test and Fisher’s exact two-tailed test analysis were performed and the differences were considered significant at values of P<0.05. Distribution of H. pylori genotypes isolated from gastric biopsy samples and also their antibiotic resistance pattern were statistically analyzed.
Ethical approval
The study was approved by the Ethical Council of Research of the Molecular Biology Research Center, Systems Biology and Poisonings Institute, Baqiyatallah University of Medical Sciences, Tehran, Iran (Consent Ref. Number BMSU-2017) (this study was conducted in accordance with the Declaration of Helsinki). Verification of this research project and the licenses related to the sampling process were approved by the Prof Reza Ranjbar (Approval Ref. Number 2017/33). Samples were collected from volunteer patients who were referred to the hospital for routine checkup. Written informed consent was obtained from all patients or their parents. Name, surname and other personal information of patients of the present study were kept secret. Additionally, we confirm that all methods were performed in accordance with the relevant guidelines and regulations.
Results
Clinical isolates
Totally, 112 ruminants, 19 children and 507 adult human gastric biopsy samples were studied for the presence of H. pylori. No relationship was found between the prevalence of H. pylori infection and the age of patients ranging from 3 to 72 years. Figure 1 represents the prevalence of H. pylori strains isolated from different types of gastric biopsy samples. We found that the prevalence of H. pylori strains in gastric biopsy samples studied using the urease test, culture and histological examination was 57.04%, 55.40% and 60.80%, respectively. Based on the culture method, samples which were collected from adult humans (78%) and sheep (70%) had the highest prevalence of H. pylori strains, while those of goats (0.6%) and cows (4%) had the lowest. There were no statistically significant differences between three different diagnostic methods. Findings of the culture method were confirmed using the PCR-based amplification of 16S rRNA. Table 2 represents the prevalence of H. pylori strains among different pathologic lesions of studied human. Distribution of gastric ulcers, duodenal ulcers and chronic gastritis among 700 human gastric biopsy samples were 10.40%, 15.70% and 96.50%, respectively. Distribution of gastric cancer and chronic cancer were 0.60% and 3.14%, respectively.
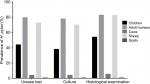  | Figure 1 Prevalence of H. pylori infection determined by diagnostic tests. |
  | Table 2 Prevalence of H. pylori infection in patients (adult humans and children) |
Antimicrobial resistance pattern
Table 3 represents the antibiotic resistance pattern of H. pylori strains isolated from different types of gastric biopsy samples. We found that H. pylori strains harbored the highest prevalence of resistance against ampicillin (74.4%), clarithromycin (63.4%), trimethoprim (61.5%), metronidazole (61.5%), erythromycin (60.1%) and amoxicillin (60.1%). Prevalence of resistance against cefsulodin, furazolidone and moxifloxacin was low. Statistically significant differences were found between the type of samples and prevalence of antibiotic resistance (P<0.05).
Genotyping pattern
Table 4 represents the distribution of different genotypes among the H. pylori strains isolated from different types of gastric biopsy samples. Total distribution of cagE, cagA, iceA1, iceA2, oipA and babA2 virulence factors was 48.5%, 87.3%, 47%, 47%, 15.2% and 27.4%, respectively. We found that the most commonly detected genotypes among the H. pylori strains isolated from different types of biopsy samples were cagA (84.79%), vacA m2 (55.95%), vacA s1a (49.84%), cagE (48.58%), iceA1 (47.02%) and iceA2 (47.02%). Prevalence of vacA mib (6.58%), oipA (15.20%) and vacA s2 (14.42%) was low. Statistically significant differences were found between the type of samples and prevalence of different genotypes (P<0.05).
  | Table 4 Total distribution of various genotypes in H. pylori strains of positive samples |
Discussion
H. pylori is an opportunistic and infectious bacterium in both humans and animals. It is also a causative agent of gastritis, duodenal ulcers, gastric ulcer and cancer in both humans and animals.23,24 However, studies conducted rarely in this field on animal cases,5–7 and all of them have demonstrated the role of the H. pylori strains as a causative agent of gastrointestinal diseases in animals.23–26 High prevalence of H. pylori strains in different types of foods with animal origins showed that animals and especially ruminants may play an important role in the maintenance and transmission of infection to humans.
As far as we know, the present study is one of the most comprehensive reports of prevalence and genotype of the H. pylori strains isolated from both humans and animals. Our findings showed that the prevalence of H. pylori strains in gastric biopsy samples of cows, sheep and goats was 4%, 70% and 0.66%, respectively. Prevalence of H. pylori strains in gastric ulcer, duodenal ulcer, gastric cancer, chronic gastritis and chronic cancer of adults and children was 10.1% and 10.5%, 15.2% and 15.7%, 0.62% and 0%, 94.1% and 89.4% and finally 2.28% and 31.5%, respectively. High prevalence of H. pylori in our study maybe due to the fact that Iran is one of the main sites of H. pylori infections in the world and therefore, pathogenic bacteria can easily transmit from infected animals or humans to healthy ones. High prevalence of H. pylori infection has also been reported in Iran.27,28 Another important reason is the fact that all samples were taken from the sites of H. pylori infection and the distribution of H. pylori strains is higher than other bacteria that exist in the site of the gastric ulcer, duodenal ulcer, gastric cancer, chronic gastritis and chronic cancer in both humans and animals.26,29–31 There were also many studies that showed the higher prevalence of H. pylori in the sites of gastrointestinal disorders in both humans and animals.32,33 Another important reason maybe the high prevalence of H. pylori strains in several types of food samples reported in previous Iranian researches.5,6,34,35 Therefore, it is not surprising that the H. pylori strains can easily transmit from the foods to humans and animals.
The prevalence of H. pylori in our study was around 40%–80% which is similar to previous reports from Iran, South America, Japan, Turkey and Pakistan, where more than 80% of dyspepsia patients were H. pylori positive; however, in Scandinavia and England, the prevalence ranges between 20% and 40%.36,37 The diversity of H. pylori frequency in numerous hosts and regions may relate to animal, microbe and environmental factors.
Another part of our investigation focused on the prevalence of antibiotic resistance in the H. pylori strains isolated from animals and humans. We found that bacterial strains harbored a high prevalence of resistance against commonly used antibiotics in the field of H. pylori infections. Prescription of ampicillin, amoxicillin, metronidazole, erythromycin and clarithromycin is common in the treatment of cases of H. pylori infections in Iran. Therefore, it is not surprising that the majority of bacterial strains were resistant against these types of antibiotics. Unauthorized and irregular prescription of antibiotics and also self-treatment of infections by patients are the main factors that affected the high prevalence of antibiotic resistance. Another Iranian study38 reported the high prevalence of resistance of H. pylori strains against ampicillin, amoxicillin, metronidazole, erythromycin and clarithromycin. Recent meta-analysis study39 showed that the rates of H. pylori antibiotic resistance reported from Asian, American, African and European countries were 47.22% (30.5%–75.02%) for metronidazole, 19.74% (5.46%–30.8%) for clarithromycin, 18.94% (14.19%–25.28%) for levofloxacin, 14.67% (2%–40.87%) for amoxicillin, 11.70% (0%–50%) for tetracycline, 11.5% (0%–23%) for furazolidone and 6.75% (1%–12.45%) for rifabutin. Because clarithromycin is the most potent antibiotic involved in the management of H. pylori infections, resistance to clarithromycin is important. We found that H. pylori strains showed high prevalence of resistance against clarithromycin (61%–100%). This prevalence of resistance against clarithromycin was higher than those in Norway (5.9%), Spain (32.01%) and Portugal (42.35%).39 Besides, the prevalence rate was also higher than India (58.8%), China (46.54%) and Malaysia (2.4%).39 This finding is due to the higher prescription of the macrolides in Iran. In recent years, due to extensive prescription of clarithromycin especially for the infections of the respiratory tracts especially in children, clarithromycin resistance has increased in various regions.38,39 Metronidazole is used to treat H. pylori infections and is one of the few antibacterial agents as a drug of choice that is effective in eradicating the H. pylori. In keeping with this, this antibiotic is used for the treatment of transient gastrointestinal and anaerobic infections in Iran. The rate of self-prescription and also self-treatment with this antibiotic is high among the Iranian patients. It is also used for the treatment of gynecological, dental and parasitic-related infectious diseases. Therefore, it is not surprising that majority of H. pylori strains were resistant against this antibiotic agent (59%–100%). Literature-based reports showed that metronidazole resistance is the most common antibiotic resistance in H. pylori and overall metronidazole resistance found in 47.22% in descending order in Africa 75.02%, South America 52.85%, Asia 46.57%, Europe 31.19%, to 30.5% in North America.39 Amoxicillin is another important choice drug for treatment of H. pylori infections in Iran. It is mainly used for the treatment of H. pylori infections in regions with high prevalence of resistance against metronidazole. We found that the prevalence of resistance against amoxicillin was lower than other tested antibiotics (10%–24%). The prevalence of amoxicillin resistance in American and European countries is low from zero in Norway, Germany, Finland and Poland, 1.4% in Spain to 2% in the United States.16 However, the prevalence of resistance against this antibiotic in South Nigeria, Colombia, India and Africa was 66%, 20.5%, 72.5% and 97.5%, respectively.39
Another part of our study focused on the detection of putative genotypes of the H. pylori strains. We found that H. pylori strains isolated from human clinical samples harbored the higher prevalence of vacA, cagA, iceA, oipA and babA genotypes. However, the prevalence of these genotypes among the animal-based strains was also high. Prevalence of cagE genotype in H. pylori strains recovered from animals was higher than humans. H. pylori strains isolated from the biopsy samples of goats harbored the highest prevalence of studied genotypes. Similar profile of the genotypes among the H. pylori strains isolated from humans and animals maybe due to the zoonotic aspect of the bacteria. There were several studies that reported the high prevalence of isolation of H. pylori from foods with animal origins.5,6,35,40,41 Therefore, it is not far from the mind that the H. pylori strains isolated from humans and animals of our study have common primary origins or have been transmitted from animals to humans or vice versa. Detection of mentioned genotypes in animals shows that the H. pylori strains may have significant roles in occurrence of gastrointestinal diseases and disorders in cows, sheep and goats.
There was only one research about the comparison of the prevalence and genotype pattern of the H. pylori strains between humans and animals.26 This study showed that overall 6 (3%) cows, 32 (16%) sheep and 164 (82%) human being specimens were positive for H. pylori strains. The vacA s1a/m1a was the predominant H. pylori genotype in cow, sheep and human samples. There was 3.4%–8.4% variability and 92.9%–98.5% homology between the genotypes of the H. pylori strains isolated from sheep and human samples. Another study42 that was conducted on food samples reported that the most commonly detected genotypes were vacA s1a (78.37%), vacA m2(75.67%), vacA m1a (51.35%) and cagA (41.89%). They showed that the prevalence of iceA1, iceA2 and oipA genotypes was 13.51%, 4.05% and 18.91%, respectively, which was lower than our findings.
The geographic distribution of different H. pylori genotypes remains mainly unknown. Prevalence of virulent genotypes in Europe and America may have significant epidemiological consequences that are linked to the presence of the cagA gene and the severity of H. pylori-related diseases.43 CagA is the most extensively studied H. pylori virulence factor. Prevalence of cagA-positive strains in Europe is around 60%–70%, while the prevalence in Asian countries is 90%.43 Such differences in the prevalence of cagA genotypes could not be clarified exactly; however, they have been related to the genetic heterogeneity or to differences in the geographic locations and maybe differences in the food habits. Documented data reported that cagA genotypes are the main factors responsible for the occurrence of gastric cancer and other malignant neoplasms in animal models.20 Since it has been demonstrated that H. pylori carries only a single copy of the vacA gene, detection of multiple genotypes implies the presence of multiple strains in a clinical sample. We found that the vacA s1a and m2 were predominant genotypes in the H. pylori strains. These genotypes are accompanied by the occurrence of human clinical and gastrointestinal diseases.44,45 VacA is the second most widely studied H. pylori virulence factor. It can induce multiple cellular activities, including membrane channel formation, cytochrome c release from mitochondria leading to apoptosis and binding to cell-membrane receptors followed by initiation of a proinflammatory response.20
Conclusion
Total prevalence of H. pylori infection in the Chaharmahal va Bakhtiari Province, Iran, was high. Virulence factors (vacA, cagA, oipA, cagE, BabA2 and iceA) of H. pylori in Iran might contribute to the low rate of gastric cancer. However, the presence of cagA is associated with more severe clinical outcomes of gastroduodenal diseases. High prevalence of virulent and resistant H. pylori strains shows a significant public health problem. Similarity in the antibiotic resistance pattern and also distribution of different genotypes in the H. pylori strains of human and animal sources may show that they had similar routes of infections. Additionally, similar antibiotic resistance and genotyping pattern may show that animals and especially ruminants are the probable sources for transmission of H. pylori infections to human population. However, additional studies are required to determine the exact epidemiological aspects of H. pylori infections in humans and animals.
Acknowledgments
We would like to thank the “Clinical Research Development Center of Baqiyatallah Hospital” for their kind cooperation. This study was financially supported in part by “Clinical Research Development Center of Baqiyatallah Hospital”. The authors also gratefully acknowledge all the staff of Biotechnology Research Center, Shahr-e Kord Branch, Islamic Azad University, Shahr-e Kord in southwest Iran for their sincere support.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Disclosure
The authors report no conflicts of interest in this work.
References
Hooi JKY, Lai WY, Wk N. Global prevalence of Helicobacter pylori infection. Gastroenterology. 2017:1–10. | ||
Sjökvist Ottsjö L, Jeverstam F, Yrlid L, Wenzel AU, Walduck AK, Raghavan S. Induction of mucosal immune responses against Helicobacter pylori infection after sublingual and intragastric route of immunization. Immunology. 2017;150(2):172–183. | ||
Bagheri N, Azadegan-Dehkordi F, Rafieian-Kopaei M, Rahimian G, Asadi-Samani M, Shirzad H. Clinical relevance of Helicobacter pylori virulence factors in Iranian patients with gastrointestinal diseases. Microb Pathog. 2016;100:154–162. | ||
Bagheri N, Azadegan-Dehkordi F, Rahimian G, Rafieian-Kopaei M, Shirzad H. Role of regulatory T-cells in different clinical expressions of Helicobacter pylori infection. Arch Med Res. 2016;47(4):245–254. | ||
Atapoor S, Safarpoor Dehkordi F, Rahimi E. Detection of helicobacter pylori in various types of vegetables and salads. Jundishapur J Microbiol. 2014;7(5):e10013. | ||
Mousavi S, Dehkordi FS, Rahimi E. Virulence factors and antibiotic resistance of Helicobacter pylori isolated from raw milk and unpasteurized dairy products in Iran. J Venom Anim Toxins Incl Trop Dis. 2014;20:51. | ||
Yahaghi E, Khamesipour F, Mashayekhi F, et al. Helicobacter pylori in vegetables and salads: genotyping and antimicrobial resistance properties. Biomed Res Int. 2014;2014:757941. | ||
Ranjbar R, Bolkheir A, Vahdat K, Assadi M, Darabi H, Nabipour I. The association of Chlamydia pneumonia and Helicobacter pylori IgG seropositivity with Omentin-1, visfatin and adiponectin levels in postmenopausal women. Acta Med Iran. 2016;54(12):771–777. | ||
O’Connor A, O’Morain CA, Ford AC. Population screening and treatment of Helicobacter pylori infection. Nat Rev Gastroenterol Hepatol. 2017;14(4):230–240. | ||
Brawner KM, Kumar R, Serrano CA. Helicobacter pylori infection is associated with an altered gastric microbiota in children. Mucosal Immunol. 2017;10(5):1169–1177. | ||
Awdalla HI, Ragab MH, Hanna LN. Environmental risk factors affecting transmission of Helicobacter pylori infection in Egypt. J Public Health. 2010;18(3):237–244. | ||
Izadi M, Fazel M, Karbasi-Afshar R, et al. Glycemic control in type 2 diabetes mellitus prevents coronary arterial wall infection. ARYA Atheroscler. 2014;10(3):141–146. | ||
Ranjbar R, Khamesipour F, Jonaidi-Jafari N, Rahimi E. Helicobacter pylori isolated from Iranian drinking water: vacA, cagA, iceA, oipA and babA2 genotype status and antimicrobial resistance properties. FEBS Open Bio. 2016;6(5):433–441. | ||
Maeda M, Moro H, Ushijima T. Mechanisms for the induction of gastric cancer by Helicobacter pylori infection: aberrant DNA methylation pathway. Gastric Cancer. 2017;20(Suppl 1):8–15. | ||
Wallden K, Rivera-Calzada A, Waksman G. Type IV secretion systems: versatility and diversity in function. Cell Microbiol. 2010;12(9):1203–1212. | ||
Ghasemi A, Shirazi MH, Ranjbar R, Khorramizadeh MR, Daryani NE, Hosseini M. The prevalence of cagA and cagE genes in Helicobacter pylori strains isolated from different patient groups by polymerase chain reaction. Pak J Biol Sci. 2008;11(22):2579–2583. | ||
Hosseinzadeh M, Khosravi A, Saki K, Ranjbar R. Evaluation of Helicobacter pylori infection in patients with common migraine headache. Arch Med Sci. 2011;7(5):844–849. | ||
Pakbaz Z, Shirazi MH, Ranjbar R, et al. Frequency of sabA gene in Helicobacter pylori strains isolated from patients in Tehran, Iran. Iran Red Crescent Med J. 2013;15(9):767–770. | ||
Souod N, Kargar M, Doosti A, Ranjbar R, Sarshar M. Genetic analysis of cagA and vacA genes in Helicobacter pylori isolates and their relationship with gastroduodenal diseases in the west of Iran. Iran Red Crescent Med J. 2013;15(5):371–376. | ||
Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol. 2010;7(11):629–641. | ||
Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Q Rev Biol. 2001;76(3):348–349. | ||
Jorgensen JH, Turnidge JD. Susceptibility test methods: dilution and disk diffusion methods. In: Manual of Clinical Microbiology. 11th ed. Washington: American Society of Microbiology. 2015:1253–1273. | ||
Peek RM. Helicobacter pylori infection and disease: from humans to animal models. Dis Model Mech. 2008;1(1):50–55. | ||
Haesebrouck F, Pasmans F, Flahou B, et al. Gastric Helicobacters in domestic animals and nonhuman primates and their significance for human health. Clin Microbiol Rev. 2009;22(2):202–223. | ||
Safaei HG, Rahimi E, Zandi A, Rashidipour A. Helicobacter pylori as a zoonotic infection: the detection of H. pylori antigens in the milk and faeces of cows. J Res Med Sci. 2011;16(2):184–187. | ||
Momtaz H, Dabiri H, Souod N, Gholami M. Study of Helicobacter pylori genotype status in cows, sheep, goats and human beings. BMC Gastroenterol. 2014;14(1):61. | ||
Eshraghian A. Epidemiology of Helicobacter pylori infection among the healthy population in Iran and countries of the Eastern Mediterranean Region: a systematic review of prevalence and risk factors. World J Gastroenterol. 2014;20(46):17618–17625. | ||
Moosazadeh M, Lankarani KB, Afshari M. Meta-analysis of the prevalence of Helicobacter pylori infection among children and adults of Iran. Int J Prev Med. 2016;7:48. | ||
Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19(3):449–490. | ||
Bauer B, Meyer TF. The human gastric pathogen Helicobacter pylori and its association with gastric cancer and ulcer disease. Ulcers. 201;2011:11–23. | ||
Mladenova-Hristova I, Grekova O, Patel A. Zoonotic potential of Helicobacter spp. J Microbiol Immunol Infect. 2017;50(3):265–269. | ||
Mitipat N, Siripermpool P, Jadwattanakul T. The prevalence of Helicobacter pylori infection in patients with gastrointestinal symptoms in Chon Buri, Thailand. Southeast Asian J Trop Med Public Heal. 2005;36(2):341–346. | ||
Adlekha S, Chadha T, Krishnan P, Sumangala B. Prevalence of Helicobacter pylori infection among patients undergoing upper gastrointestinal endoscopy in a medical college hospital in Kerala, India. Ann Med Health Sci Res. 2013;3(4):559–563. | ||
Dimola S, Caruso ML. Helicobacter pylori in animals affecting the human habitat through the food chain. Anticancer Res. 1999;19(5B):3889–3894. | ||
Ghorbani F, Gheisari E, Dehkordi FS. Genotyping of vacA alleles of Helicobacter pylori strains. 2016;15(August):1631–1636. | ||
Javed M, Amin K, Muhammad D, Hussain A, Mahmood N. Prevalence of H. pylori. Prof Med Sep. 2010;17:431–439. | ||
Hunt RH, Xiao SD, Megraud F, et al. Helicobacter pylori in developing countries. World Gastroenterology Organisation Global Guideline. J Gastrointestin Liver Dis. 2011;20(3):299–304. | ||
Mahmoudi S, Mamishi S, Banar M, et al. Antibiotic susceptibility of Helicobacter pylori strains isolated from Iranian children: High frequency of A2143G point mutation associated with clarithromycin resistance. J Glob Antimicrob Resist. 2017;10:131–135. | ||
Ghotaslou R, Leylabadlo HE, Asl YM. Prevalence of antibiotic resistance in Helicobacter pylori: A recent literature review. World J Methodol. 2015;5(3):164–174. | ||
Gilani A, Razavilar V, Rokni N. VacA and cagA genotypes of Helicobacter pylori isolated from raw meat in Isfahan province, Iran. Vet Res forum an Int Q J. 2017;8(1):75–80. | ||
Saeidi E, Sheikhshahrokh A. vacA genotype status of Helicobacter pylori isolated from foods with animal origin. Biomed Res Int. 2016;2016:1–6. | ||
Hemmatinezhad B, Momtaz H, Rahimi E. VacA, cagA, iceA and oipA genotypes status and antimicrobial resistance properties of Helicobacter pylori isolated from various types of ready to eat foods. Ann Clin Microbiol Antimicrob. 2016;15(1):2. | ||
Saribasak H, Salih BA, Yamaoka Y, Sander E. Analysis of Helicobacter pylori genotypes and correlation with clinical outcome in Turkey. J Clin Microbiol. 2004;42(4):1648–1651. | ||
Jafari F, Shokrzadeh L, Dabiri H, et al. vacA genotypes of Helicobacter pylori in relation to cagA status and clinical outcomes in Iranian populations. Jpn J Infect Dis. 2008;61(4):290–293. | ||
Park SM, Park J, Kim JG, Yoo BC. Relevance of vacA genotypes of Helicobacter pylori to cagA status and its clinical outcome. Korean J Intern Med. 2001;16(1):8–13. | ||
Ranjbar R, Khamesipour F, Jonaidi-Jafari N, Rahimi E. Helicobacter pylori in bottled mineral water: genotyping and antimicrobial resistance properties. BMC Microbiol. 2016;16:1–10. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

