Back to Journals » Infection and Drug Resistance » Volume 12
Genetic variation in metronidazole metabolism and oxidative stress pathways in clinical Giardia lamblia assemblage A and B isolates
Authors Saghaug CS , Klotz C, Kallio JP , Brattbakk HR, Stokowy T , Aebischer T , Kursula I , Langeland N , Hanevik K
Received 25 October 2018
Accepted for publication 16 January 2019
Published 10 May 2019 Volume 2019:12 Pages 1221—1235
DOI https://doi.org/10.2147/IDR.S177997
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Christina S Saghaug, 1, 2 Christian Klotz, 3 Juha P Kallio, 4 Hans-Richard Brattbakk, 1, 5 Tomasz Stokowy, 1, 5 Toni Aebischer, 3 Inari Kursula, 4, 6 Nina Langeland, 1– 2, 7 Kurt Hanevik 1, 2
1Department of Clinical Science, University of Bergen, Bergen, Hordaland, Norway; 2Norwegian National Advisory Unit on Tropical Infectious Diseases, Department of Medicine, Haukeland University Hospital, Bergen, Hordaland, Norway; 3Department of Infectious Diseases, Unit 16 Mycotic and Parasitic Agents and Mycobacteria, Robert Koch-Institute, Berlin, Germany; 4Department of Biomedicine, University of Bergen, Bergen, Hordaland, Norway; 5Center for Medical Genetics and Molecular Medicine, Haukeland University Hospital, Bergen, Hordaland, Norway; 6Biocenter Oulu and Faculty of Biochemistry and Molecular Medicine, University of Oulu, Oulu, Finland; 7Department of Medicine, Haraldsplass Deaconess Hospital, Bergen, Hordaland, Norway
Purpose: Treatment-refractory Giardia cases have increased rapidly within the last decade. No markers of resistance nor a standardized susceptibility test have been established yet, but several enzymes and their pathways have been associated with metronidazole (MTZ) resistant Giardia. Very limited data are available regarding genetic variation in these pathways. We aimed to investigate genetic variation in metabolic pathway genes proposed to be involved in MTZ resistance in recently acquired, cultured clinical isolates.
Methods: Whole genome sequencing of 12 assemblage A2 and 8 assemblage B isolates was done, to decipher genomic variation in Giardia. Twenty-nine genes were identified in a literature search and investigated for their single nucleotide variants (SNVs) in the coding/non-coding regions of the genes, either as amino acid changing (non-synonymous SNVs) or non-changing SNVs (synonymous).
Results: In Giardia assemblage B, several genes involved in MTZ activation or oxidative stress management were found to have higher numbers of non-synonymous SNVs (thioredoxin peroxidase, nitroreductase 1, ferredoxin 2, NADH oxidase, nitroreductase 2, alcohol dehydrogenase, ferredoxin 4 and ferredoxin 1) than the average variation. For Giardia assemblage A2, the highest genetic variability was found in the ferredoxin 2, ferredoxin 6 and in nicotinamide adenine dinucleotide phosphate (NADPH) oxidoreductase putative genes. SNVs found in the ferredoxins and nitroreductases were analyzed further by alignment and homology modeling. SNVs close to the iron-sulfur cluster binding sites in nitroreductase-1 and 2 and ferredoxin 2 and 4 could potentially affect protein function. Flavohemoprotein seems to be a variable-copy gene, due to higher, but variable coverage compared to other genes investigated.
Conclusion: In clinical Giardia isolates, genetic variability is common in important genes in the MTZ metabolizing pathway and in the management of oxidative and nitrosative stress and includes high numbers of non-synonymous SNVs. Some of the identified amino acid changes could potentially affect the respective proteins important in the MTZ metabolism.
Keywords: drug metabolism, resistance, genetic analysis, metronidazole genes, ferredoxin, genetic diversity
Corrigendum for this paper has been published
Introduction
Giardia lamblia is a microaerophilic, eukaryotic and tetraploid parasite, annually responsible for up to ~280 million human infections worldwide.1 Giardia is endemic in low- and middle-income countries (LMIC), and infection with the parasite has been shown to affect growth and cognitive functions in children in resource-poor regions.2,3 In high-income countries, Giardia is usually transmitted by waterborne outbreaks or found in returning travelers from LMIC.4,5 Giardiasis is clinically associated with prolonged diarrhea and stomach pain, and it may lead to post-infectious sequela, eg, irritable bowel syndrome and chronic fatigue.6,7
The antibiotic metronidazole (MTZ) is a prodrug belonging to the 5-nitromidazole chemical group, and a common first-line treatment for giardiasis.8 With its activity against microaerophilic and anaerobic pathogens, MTZ has a treatment efficacy of 73–100%.9 MTZ enters the parasite through passive diffusion, it is activated through enzymatic reduction and releases toxic intermediates. This reaction can only occur in an anaerobic or microaerophilic environment.10,11 Effects of activated MTZ on susceptible Giardia include protein malfunction, multiplication disruption, DNA damage and oxidative stress which will lead to cell death.12–15 MTZ can be activated by four main enzymes in Giardia: pyruvate-flavodoxin oxidoreductases (PFOR-1 and 2), the thiol-cycling associated enzyme thioredoxin reductase (TrxR)16 and nitroreductase (NR)-1. The iron-sulfur (Fe-S) cluster binding proteins, ferredoxins (Fd) have also been shown to participate in activation of MTZ in Giardia.16 NR-2, a paralog of NR-1, is hypothesized to detoxify MTZ by fully reducing the drug without making toxic intermediates.17–19 While Giardia is exposed to MTZ, it also has to constantly detoxify oxygen, nitric oxide (NO) and other harmful chemicals that may affect growth and viability.20
The majority of proposed resistance mechanisms consist of either down-regulation of enzymes that render MTZ cytotoxic (NR-1, PFORs and TrxR), or up-regulation of enzymes that can make MTZ an inert molecule (NR-2).19 Most work regarding drug resistance mechanisms in Giardia has been done using laboratory induced resistant assemblage A1 isolates, which rarely infects humans, compared to infections with Giardia assemblage A2 and B, which are common. MTZ is probably activated through several pathways and exhibit a pleiotropic mode of action, and resistance to MTZ has generally taken longer time to develop compared to resistance against most other antibiotics.21
MTZ resistance in Giardia is a growing problem. Clinical experience and recent studies of returning travelers have shown that MTZ-refractory Giardia cases have a prevalence of ~20%.22,23 The prevalence has dramatically increased over the last few years, reaching up to 40% in a referral outpatient clinic in London.24 The highest prevalence of MTZ refractory cases have been found in returning travelers from Asia, especially India, or the south and east Mediterranean areas.22,24–26
No standardized susceptibility assays for Giardia isolates are available,27 and susceptibility testing in the laboratory is challenging due to genomic complexity of the parasite. Moreover, isolates are difficult to culture, and growth rates may differ between isolates and assemblages. The microaerophilic environment in which Giardia thrives is difficult to mimic in vitro for drug susceptibility assays. Experiments may be performed in unfavorable high levels of O2, affecting growth rates and MTZ metabolism of Giardia. Thus, a reliable evaluation of MTZ’s activation and tolerance can be difficult to obtain.28 Studies of MTZ metabolism pathway genes in Giardia are mostly based on gene expression analysis in isogenic laboratory-derived resistant Giardia of the assemblage AI strains, rarely found in humans.13,18,29–34 So far, no specific markers for resistance in Giardia have been discovered, and resistance is likely complex and based on several factors. These factors could include amino acid variations in proteins and altered function, post- or pre-translational variants, changes in expression of activating or protective proteins, modifications of regulatory pathways and epigenetic modifications,16,35–39 and a recent study of resistant Giardia strains showed that MTZ is capable of inducing significant changes in proteins found in the antioxidant, electron transport, and pyruvate catabolism networks. In addition, a correlation between acetylation of these proteins and MTZ resistance was found.35
Begaydarova et al 2015 investigated clinical isolates and linked PFOR gene expression to MTZ resistance, but PFOR was not found to be a good resistance marker.40 Another study by Galeh et al 201641 linked non-synonymous single nucleotide variants (nsSNVs) in PFOR and NR genes to MTZ resistance and possibly through reduced translation and thereby reduced MTZ activation. Genetic variation in Giardia genome is not well known, except for a few genotyping genes, ie, triose phosphate isomerase (tpi), glutamate dehydrogenase (gdh) and beta-giardin (bg).42 The genetic variation among Giardia isolates has in one study been found to be low for some assemblage A genes, ie, Fd3 (DHA2_154390) with more SNPs in assemblage B.43 However, the background genetic variability of the parasites’ MTZ metabolizing pathways and management of the oxidative and nitrosative stress has not been investigated in detail and constitutes an important basis for further research into potential markers of MTZ resistance.
Laboratory-induced MTZ resistance may be lost during one en/excystation cycle,44 indicating that metabolic adaptations are likely to contribute to resistance. Still, the recent emergence of rapidly increasing clinical MTZ resistance raises the suspicion that strains with a genetically inheritable trait for resistance are now circulating. Indeed, one laboratory-induced resistance line has been shown to possess a non-sense mutation in one of its NR-1 alleles.16 However, this challenge may best be studied by examining current clinical isolates rather than laboratory strains obtained decades ago. The prospect for whole genome sequencing of purified cysts from clinical stool samples makes it possible to perform such studies.45 The aim of our study was to investigate the general genetic variation in proteins involved in metabolic pathways associated with MTZ resistance or management of oxidative stress substances.
Material and methods
Genes of interest
A PubMed literature search was performed using the word Giardia in combinations with one or two of the words: MTZ, refractory, resistance and oxidative stress, to identify genes in the MTZ metabolism pathway and oxidative and nitrosative stress management in Giardia. The search aimed to identify genes that were upregulated or downregulated during exposure to MTZ and/or free radicals or associated with the activation or inactivation of MTZ and genes with antioxidant properties. These publications and their references were used to establish a list of 29 candidate genes (see Table 1). In order to identify pairwise shared genes and the correct gene ID for Giardia assemblage A2 and B, supplementary data from Adam et al was used.46 Finally, the Giardia DB (
 | Table 1 Genes included in the SNV analysis for metronidazole metabolism, oxidative- and nitrosative-stress management. The ferredoxins have been named according to Ansell et al 2017,16 and another Fd, DHA2_153401/GSB_151614, earlier presented by Nixon et al48, has been given the name Fd6 in this table. The other genes of interest have been named according to annotations from Giardia DB |
Giardia isolates
Twelve isolates of Giardia assemblage A2 and eight isolates of assemblage B were cultured from human clinical stool samples between 2011 and 2015 at the Robert Koch Institute, Berlin, Germany. The stool samples were collected by the Institute of Tropical Medicine and International Health, Charité, Berlin, Germany. One of the isolates, P324, was obtained from the Tropenmedizinische Ambulanz, University hospital Düsseldorf, Germany. No clinical data for the samples have been collected. Giardia cysts from the stool samples were purified using a two-step sucrose gradient centrifugation. The cysts were excysted using a standard two-step excystation method,49 in order to acquire trophozoites. The trophozoites were grown in Diamond’s TY-S-33 medium supplemented with bile at 37°C according to.50 Collection of the trophozoites was done from confluent cultures in 11 mL tubes, and then the DNA was extracted using Maxwell® 16 FFPE Plus LEV DNA Purification Kit (Promega BioTech AB, Sweden, cat AS1135) together with Maxwell® 16 instrument (AS1000). The quality and amount of DNA was validated with NanoDrop and the concentration of the DNA was verified with Qubit 2.0 (Life Technologies, Grand Island, NY).
Genome sequencing
Fragmented Giardia DNA (2 μg, 550 bp insert size) from the trophozoites was first prepared and indexed using Illumina TruSeq DNA PCR-Free Sample Prep Kit (Cat. Nr 20015962 and 20015960S). Library size distribution was validated using the Agilent Technologies 2100 BioAnalyzer, High Sensitivity DNA chip, and quantification was done using the KAPA Library Quantification Kit for Illumina sequencing platforms. Normalized whole genome Giardia DNA-libraries were sequenced using the Illumina MiSeq paired-end technology (2×300 bp). FASTQ-files from the sequencing were assembled and aligned to the corresponding reference genomes of Giardia assemblage A2 and B (v.26) using Bowtie 2-2.2.3.51 The reference genomes were obtained from Giardia DB in January 2016 versions 2013-11-25.52
Variant analysis
Aligned data were analyzed using Geneious v.10.2.4. Variant calling was done to identify SNVs using the parameters minimum variant frequency 0.1 and a minimum coverage of 10 nucleotides. The coding regions (CDS) of 29 candidate genes (including pre, 150 bp, - and post-coding regions, 50 bp) were extracted from Giardia assemblage A2 and B in Geneious. For genes with introns (Fd3 in both assemblages), genetic analysis was done in the CDS and SNVs in introns are therefore not presented. To obtain all the SNVs in one alignment, the 29 genes from each isolate were concatenated, and then compared against each other assemblage-wise. Comparisons of the 29 genes were also performed in Geneious using a global alignment with free end gaps. Alignments of ferredoxin domains are based on findings from Nixon et al 2002,48 and for more detailed information see the
Homology modeling of the nitroreductases and ferredoxins
Homology modeling of the NRs and ferredoxins 1–6 were done based on the amino acid sequences used in the gene analysis. Models were created using Protein Homology/analogY Recognition Engine V 2.0 (Phyre2) server with intensive mode.53 Models were visualized and superimposed in PyMol (The PyMOL Molecular Graphics System, Version 2.1.0 Schrödinger, LLC).
Storage of data
All genetic data is kept on a secure server; Norwegian e-infrastructure for Life Sciences.
Results
Whole genome sequencing of 20 cultured Giardia assemblage A2 and B isolates was successfully carried out with an average coverage of 31.1 (110.3) (see
Based on the criteria in the literature search, 29 candidate genes were selected and are presented in Table 1.
For the 29 candidate genes, the genetic variation was analyzed across the 12 assemblage A2 and eight assemblage B clinical isolates. The average number of nsSNVs per gene length was 0.3 % for assemblage A2 and 1.2% for assemblage B. Averages for SNVs for each gene are presented in
Genes involved in the MTZ and oxidative stress and nitrosative stress management
Considerably more nsSNVs could be found in the Giardia assemblage B isolates than in the assemblage A2 isolates. A color-graded heatmap of the nsSNVs causing amino acid changes in the 29 proteins involved in MTZ metabolism, oxidative and nitrosative stress management is therefore only shown for Giardia assemblage B (Figure 1). The genetic variation in the flavohemoprotein (GSB_151570) is not illustrated in Figure 1 due to high frequency of SNVs and variably higher coverage than the average.
For two of the genes with highest frequency of genetic variability in assemblage B (GSB_151294 - thioredoxin peroxidase and GSB_153178 – NR-1), the nsSNVs constituted over 4% of the nucleotides in the gene, and only two genes were found to be without any nsSNVs (histone H2A; GSB_151412 and Fd6; GSB_151614). The average percentage of any SNV per CDS length was 5.4% in Giardia assemblage B and 1.1% in assemblage A2.
The majority of the genes was shown to have at least one sSNV, except for the four genes Fd3; DHA2_154390, NR-1; DHA2_153380, Fd1; DHA2_9662 and NADPH oxidoreductase putative; DHA2_17151 in assemblage A2. sSNVs were present in all genes in assemblage B. SNVs in the pre-CDS regions were shown to be higher in assemblage B (average of 10.1 SNVs in 150 bp) than in assemblage A2, where the average was found to be 1.1 SNVs per 150 bps. Post-CDS regions had generally lower variability (0.3 in A2 and 4.2 in B for 50 bp).
Nitroreductases
Three different NRs were investigated for the presence of SNVs in this study, the NR-1, NR-2 and the oxygen-insensitive NTR-1. NR-1, had one of the highest numbers of nsSNVs (4.1%) for Giardia assemblage B, with more nsSNVs than sSNVs in the CDS. However, there were lower than average numbers of SNVs in the non-coding regions of the NR-1 in assemblage B. The NR-1 gene in assemblage A2 was found to be fully conserved without any SNVs in the coding or non-coding regions. The NR-2 gene sequence also had high numbers of nsSNVs in Giardia assemblage B (2.1%), and SNVs in the pre-CDS regions were higher than the average (21 vs 10.1 out of 150 bp). The NR-2 in Giardia assemblage A2 had low numbers of SNVs, where only one was a nsSNV. The NTR-1 gene was fully conserved in assemblage A2, and had lower than average numbers on non-syn SNVs in B.
Ferredoxins
The Fds are proposed to be co-factors for the MTZ activating proteins, PFOR-1 and 2, and changes in these co-factors could potentially affect their function, binding and role. We found that three of the ferredoxins in the Giardia assemblage B isolates, Fd2; GSB_10329, Fd4; GSB_153527 and Fd1; GSB_9662, had higher numbers of nsSNVs than the average of 1.2% (respectively 3.1%, 1.7% and 1.7%). Interestingly the Fd2 was one of the proteins with the most SNVs in the post- and pre-CDS regions of all the genes investigated in assemblage B, and it also had a high number of nsSNVs in the coding region.
For Giardia assemblage A2 two of the ferredoxins had higher numbers of nsSNVs than the average of 0.3%, respectively Fd2; DHA2_10329 (2.1%) and Fd6; DHA2_153401 (1.1%). One fascinating finding was that the Fd6 was fully conserved in Giardia assemblage B while assemblage A2 had some SNVs present.
Antioxidant and metabolic proteins
Two of the free radical protective enzymes, prx (GSB_151294) and NADH oxidase (GSB_9719) and the metabolic protein alcohol dehydrogenase (GSB_13350) was demonstrated to have higher numbers of nsSNVs in assemblage B than the average. Post-CDS analysis revealed that TrxR; GSB_9827 was one of the proteins with the highest number of SNVs, while NADPH oxidoreductase putative; GSB_17150, the two prxs, GSB_153801 and GSB_14521, malate dehydrogenase; GSB_3331, flavoprotein; GSB_10358 and alcohol dehydrogenase; GSB_13350, all had higher numbers of post-CDS SNVs than the average.
Alignment of the ferredoxins and the nitroreductases
Some of the ferredoxins and NRs were found to have high numbers of nsSNVs in many of the Giardia assemblage A and B isolates. The amino acid changing SNVs and their positions in the proteins were therefore analyzed further, and an alignment was made to compare the cysteine domains in the five different ferredoxin proteins included in this analysis in addition to NR-1 and NR-2 and a hypothetical Fd previously described in WB (Fd5)48 The proteins NR1, Fd1, and Fd3 in assemblage A2 and Fd3 and Fd6 in assemblage B had no nsSNVs in their ferredoxin domains. The nsSNVs positioned around the cysteine residues in the ferredoxin domains responsible for forming the Fe-S clusters binding sites (electron transport sites) are shown in Figure 2.
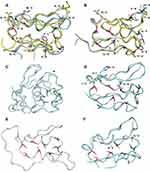
Homology modeling of the ferredoxin domains
In order to be able to analyze the effect of the amino acid changes, homology modeling was used. The ferredoxin domain sequence alignment presented in Figure 2 and the putative cysteine residues for binding Fe-S clusters are coded with the same color codes as shown in the homology models of the domains in Figure 3. Homology modeling indicates various differences within the ferredoxin domains.
Fd1, Fd2 and Fd4 can be superimposed on ferredoxin domains of NR-1 and 2. All of them clearly have two cysteine motifs, with four cysteines each, able to bind Fe-S clusters, probably of the type 4Fe-4S.55,56 The rest of the ferredoxins differ from the others by shape and ability to bind Fe-S clusters. The Fd6 has two cysteine motifs but one of the motifs only has three coordinating cysteines instead of four, which could indicate that the Fd6 only can bind one 4Fe-4S cluster or potentially one 4Fe-4S and an additional 3Fe-4S cluster (bound to 3-cystein motif). The Fd3 has only one clear motif for binding 4Fe-4S cluster and in addition it has a proposed elongated C-terminus. Fd5 has both an elongated N- and a C-terminus.
Some of the amino acid substitutions were positioned close to the ferredoxin domain with Fe-S cluster binding cysteines and sufficiently different in charge or hydrophobicity to be able to potentially cause a change significant enough to affect protein function. Some of the amino acids in the ferredoxin domains are surface exposed and changes here could potentially affect the proteins’ ability to interact with other proteins in possible intermolecular electron transfers. These changes include changes from a positively charged amino acid such as K to a negatively charged E or D seen in NR-1 assemblage B, or a change from E to K in Fd2 assemblage A2 and in Fd2 from assemblage B where G is changed to R.
In NR-1 from assemblage B, S can be changed to P (pos. 70 Figure 2), and in NR-2 from A2, P can change to S (pos. 41 in Figure 2) which might change the conformation of the protein backbone, as P is structurally more rigid than other amino acids. These SNVs are located next to or between two cysteines coordinating Fe-S cluster binding in NR-1 and NR-2, respectively. SNVs that could potentially have an effect to electrostatic properties of Fe-S clusters are G to R (pos 66) change in NR-2 from assemblage B and G to E (pos 38) change in Fd4 from assemblage B.
Discussion
The genome of Giardia trophozoites is tetraploid, consisting of two diploid nuclei. The variability among and within the assemblages can therefore potentially be high. In our study we show that the nsSNVs are common in the investigated genes in culturable clinical isolates of Giardia assemblage A2 and B. Generally, Giardia assemblage B had higher numbers of SNVs in the 29 genes investigated, and relates well to the earlier finding that the allelic sequence heterozygosity (ASH) in assemblage B is up to 0.53%,57 whereas the ASH of two A2 isolates, AS-98 and AS-175, has been determined to be 0.25–0.35%.46 In Giardia assemblage B, particularly high numbers of nsSNVs were found in gene sequences encoding MTZ activating enzyme NR-1, the PFOR co-factors Fd2, Fd1 and Fd4, the protective enzymes prx and NADH oxidase, the metabolic enzyme alcohol dehydrogenase, in addition to the MTZ inactivating enzyme NR-2. In assemblage A2, gene sequences of Fd2, Fd6 and the ROS-contributing enzyme NADPH oxidoreductase putative had the greatest number of nsSNVs.
Homology modeling shows that some of the amino acid changing SNVs are in close proximity to the cysteine residues responsible for forming Fe-S clusters and could potentially affect binding affinities of the proteins NR-1, NR-2, Fd2 and Fd4, or how they may exert their roles as electron transferring proteins.
Genetic variation as potential resistance markers
High numbers of nsSNVs found in the Giardia assemblage B isolates may indicate that assemblage B mutates faster than assemblage A2 isolates. The higher variation frequencies found in the B isolates may be associated with greater functional variation, higher prevalence of disease in humans and higher rates of treatment-refractory cases.58,59
A recent study,41 using clinical Giardia isolates, suggested that nsSNVs in NR-2 and PFOR genes could be linked to resistant Giardia isolates, but the study was based on only two treatment-refractory isolates. In addition, this study gives potential evidence that different haplotypes of these two genes may exist within the genome. This finding correlates with our observation that the numbers of nsSNVs in the NR-2 protein in Giardia assemblage B was higher than average. The PFORs in our study were found to have lower numbers of nsSNVs than average. However, the co-factors for the PFORs, Fds, were found to be highly variable within three of the five Fds in assemblage B and two of the Fds in Giardia assemblage A2.
The role of the MTZ activating enzyme, PFOR, is thought to be the transfer of electrons, through its bound Fe-S clusters, to soluble Fds.38 Lower levels of PFOR and ferredoxin enzyme activities have been characterized in resistant Giardia cultures and were also connected to MTZ sensitivity in clinical isolates.16,32,60,61 PFOR gene expression was, however, not found to be of significant difference in patient isolates with different susceptibility to MTZ in vitro.40
In other anaerobe organisms, genetic markers of resistance have been identified. The enzyme known as oxygen-insensitive nitroreductase, has been shown to be an MTZ activating enzyme in Helicobacter pylori and Escherichia coli, and bacteria with inactivated NTR or nsSNVs in this gene, may be resistant.62–64 Additionally, resistance in the parasite Trichomonas vaginalis has been associated to SNVs in the two nitroreductase genes ntr4Tv and ntr6Tv.65 SNVs found in intergenic regions of 13 genes in T. vaginalis have been linked to resistance as well, and the study suggests that a panel of genetic markers could be used as a diagnostic tool of resistance.66 Intergenic analysis should be done in Giardia as well, in order to predict whether SNVs here may play a role in MTZ resistance.
An enzyme in Giardia known as oxygen-insensitive nitroreductase (NTR-1), is similar to the NTR in bacteria, and it might have been acquired through lateral transfer from bacteria.48 The NTR-1 is different from the NR-1 and NR-2 due to missing ferredoxin domains in the protein and may not play a key role in the Fe-S cluster electron transportation. We found the presence of nsSNVs in NTR-1 to be low in our analysis. Whether this enzyme has a role in MTZ activation in Giardia is not known, but it may have a fundamental role in protection against oxidative- and nitrosative stress, as it is O2 insensitive, and has been shown to be upregulated during MTZ exposure.15
Giardia could also potentially down-regulate enzymes that detoxify oxygen to protect itself from the activated form of MTZ. This down-regulation is favorable due to the fact that MTZ may only be reduced in microaerophilic or anaerobic environments. Presence of oxygen helps to deactivate MTZ by futile cycling, creating ROS.60 The ROS will then require extensive detoxification by enzymes in the antioxidant system to avoid harmful cell damage.67 In Giardia assemblage B, high variability was found in the two oxygen detoxifying proteins NADH oxidase (GSB_9719) while the ROS contributing enzyme NADPH oxidoreductase putative (GSB_17150) was found to have higher variability than the average for both assemblages. Oxidoreductases have previously been shown to be upregulated when Giardia is challenged with H2O2.37 The functions and characteristics of the NADPH oxidoreductases and their potential paralogs should be investigated in future studies, to understand whether they have a role MTZ resistance, as oxidoreductases with null mutations, causing non-functional proteins, have been associated with MTZ resistance in H. pylori.68
Some of the genes investigated in the present study were found to have few, or non-existent, nsSNVs (Fd6 and Histone H2A in assemblage B, in addition to 10 genes in assemblage A2, see
Resistance induced in the laboratory may have different manifestations than clinical resistance, and the use of Giardia isolates directly from patients, without undergoing laboratory culturing are likely to supplement studies with isogenic laboratory strains.69,70
Non-coding regions
SNVs found in non-CDS regions of genes could affect regulatory elements and promoters which may further potentially affect gene transcription, mRNA translation and next protein expression.35,71 The genes, NR-1 and the PFOR co-factor Fd2, were found to have especially high numbers of pre-CDS SNVs in Giardia assemblage B in our study. The sequence preceding the CDS, harbor transcription promoters, and based on scarce knowledge of these in Giardia, they are short, around 50 bp long, initiator-like AT-rich sequences approximately −65 to −29 base pairs upstream the ATG start codon.57,72 Low conservation of the promoters in Giardia has previously been reported to be common, and may be related to our findings.72 How the presence of SNVs in the promoter regions of genes in Giardia may affect transcription of the genes is however not known yet. The interpretation of the SNV findings in these regions are therefore limited, and in this study, we decided to present them, but not to evaluate their potential regulatory roles.
Ferredoxin domains and SNVs
The alignment of the NRs and Fds in this study, only include the ferredoxin domains, responsible for binding the Fe-S clusters (see Figure 2). This region has earlier been associated with drug activation, as the electrons are thought to be transferred from the ferredoxin domain to the active reduction center in order to reduce MTZ.18 The active reducing center in the NRs has earlier been characterized as a nitro-flavin mononucleotide (FMN) reductase domain.18
Most of the nsSNVs present in the ferredoxin domain alignment in Figure 2 will probably not alter the function of the proteins as the changes do not alter the basic properties, eg, polarity or charge, of the amino acid residues.
Some other changes are located close to the cysteine part of the 4Fe-4S binding motif and might, therefore, affect the geometry or the electron transfer properties of the Fe-S cluster. Activated MTZ has previously been speculated to be able to react with nucleotides and cysteines (free thiol groups) and form adducts.38 Whether MTZ can form adducts with the proteins investigated in the present study and whether amino acid substitutions close to the cysteine domains could affect formation must be investigated further using structural- and molecular biology studies.
Other proteins that also have distinct Fd domains include the two large proteins PFOR-1 and PFOR-2. They have been hypothesized to have three motifs to bind 4FeS4 cluster. Even if the PFORs have their own Fd domains, they are still thought to use Fds in the cytosol for electron transfer and MTZ activation.38
Coverage and multicopy genes
Several of the genes in this study were found to have paralogs annotated in the genomes of Giardia assemblage A2 and B, and we aimed to include genes present in both of the Giardia assemblages. A total of 12 paralogs were found for the NADPH oxidoreductase, and an alignment was done to compare their similarity (data not shown). The alignment showed they were only 78.3% similar to one another, and probably not multicopy genes.
We were not able to investigate the true number of sSNVs and nsSNVs of the gene sequence of flavohemoprotein due to its variable higher coverage than the other genes, indicating that it is present in a variable number of copies in different isolates (
Two shorter paralogs of flavohemoprotein, DHA2_153759 and DHA2_152971 were identified, and had high identity to the longer flavohemoprotein DHA2_154000 (data not shown). Functions of these two proteins should be investigated in detail in the future to know if they have similar properties as the DHA2_154000, or if they potentially could be wrongly annotated or fragmented versions of DHA2_154000. The shorter flavohemoproteins could have affected the coverage of the Giardia assemblage A2 isolates, as they each would align to the 5ʹ or 3ʹ end of the DHA2_154000 gene. They do not overlap, leaving approx. 500 bp in the middle of the gene. The coverage was nevertheless even, and the two paralogs in assemblage A2 would also not explain the higher coverage of the assemblage B gene GSB_151570. The potential copy-number variation in this gene, coupled with amino acid changing SNVs could be important for understanding Giardia’s capability of handling oxidative stress, as flavohemoprotein is an important NO and O2 scavenging enzyme.16,20,36,37,73–76
We also noted that histone H2A had 24 times lower coverage than the average in one of the assemblage A2 isolates. Possibly this gene could be missing, or found in another contig in this isolate, or a sequencing error may have occurred.
WGS limitations
The Illumina MiSeq paired-end technology sequencing used in our study gave a low coverage for several of the isolates and their genes, limiting the SNV analysis. For two assemblage B and one assemblage, A2 isolates the average coverage for the genes was only 11.3–14.0 (
Conclusion
Genetic variation in the form of SNVs is common in both of the human-infecting Giardia assemblages A2 and B. Assemblage B showed higher variation than assemblage A2, and presence of amino acid changing SNVs were especially high in genes associated with MTZ metabolism (ferredoxins, nitroreductase-1 and 2) and for some of the genes important in the detoxification of free radicals (thioredoxin peroxidase and NADH oxidase). Some of the discovered SNVs in this study could potentially affect the electron transfer properties of Fe-S clusters in some of the NRs and Fds investigated. Further studies are needed to address their functional properties, but study design can be guided by the presented findings. Analysis of genetic variation in circulating MTZ susceptible and treatment-refractory clinical Giardia isolates will be important to address the recent increase in MTZ resistant Giardia cases. The present study can be regarded as a basis for further studies into how genes in the metabolizing pathways may differ between resistant and susceptible Giardia isolates.
Ethical aspects
No patient data were used in the present study and any link between individual parasite data and patient information had been removed before WGS of trophozoites was carried out.
Data availability
The dataset used for this analysis and supporting the conclusions of this article can be found in GenBank with the accession numbers: MK043361 - MK043940.
Acknowledgments
We would like to thank the personnel at the Genomics Core Facility at Oslo University Hospital (oslo.genomics.no) for whole genome sequencing of the Giardia DNA and the personnel at the Department of Clinical Science, University of Bergen, for technical assistance with the sequencing. We acknowledge Prof Dr Ralf Ignatius from the Institute of Tropical Medicine and International Health, Charité, Berlin, Germany for kindly providing biological samples. We thank Petra Gosten-Heinrich, Robert Koch-Institute Berlin, for excellent technical assistance. This study was supported by the University of Bergen, the Norwegian National Advisory Unit on Tropical Infectious Diseases, Department of Medicine, Haukeland University Hospital, Helse Vest and Norsk overvaakningssystem for antibiotikaresistens hos mikrober (NORM). All the experiments, data analysis and evaluation were performed independently by the authors, without any interference from the funding institutions.
Author contributions
Culture of Giardia isolates and DNA extraction was performed by CK. Laboratory work related to whole genome sequencing and mapping of whole genome sequences was performed by HRB. SNV identification and alignment of the genes was performed by CSS. Homology modeling and amino acid change effects analysis for protein function was done by JPK and IK. CSS drafted the manuscript and KH supervised all parts of the study. All authors contributed toward data analysis, drafting and critically revising the paper, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Lane S, Lloyd D. Current trends in research into the waterborne parasite Giardia. Crit Rev Microbiol. 2002;28(2):123–147. doi:10.1080/1040-840291046713
2. Berkman DS, Lescano AG, Gilman RH, Lopez SL, Black MM. Effects of stunting, diarrhoeal disease, and parasitic infection during infancy on cognition in late childhood: a follow-up study. Lancet (London, England). 2002;359(9306):564–571. doi:10.1016/S0140-6736(02)07744-9
3. Rogawski ET, Platts-Mills JA, Seidman JC, et al. Early antibiotic exposure in low-resource settings is associated with increased weight in the first two years of life. J Pediatr Gastroenterol Nutr. 2017;65(3):350–356.
4. Craun GF, Calderon RL, Craun MF. Outbreaks associated with recreational water in the United States. Int J Environ Health Res. 2005;15(4):243–262.
5. Farthing MJ. Giardiasis. Gastroenterol Clin North Am. 1996;25(3):493–515.
6. Hanevik K, Dizdar V, Langeland N, Hausken T. Development of functional gastrointestinal disorders after Giardia lamblia infection. BMC Gastroenterol. 2009;9:27.
7. Persson R, Wensaas KA, Hanevik K, Eide GE, Langeland N, Rortveit G. The relationship between irritable bowel syndrome, functional dyspepsia, chronic fatigue and overactive bladder syndrome: a controlled study 6 years after acute gastrointestinal infection. BMC Gastroenterol. 2015;15:66.
8. Lalle M. Giardiasis in the post genomic era: treatment, drug resistance and novel therapeutic perspectives. Infect Disord Drug Targets. 2010;10(4):283–294.
9. Solaymani-Mohammadi S, Genkinger JM, Loffredo CA, Singer SM. A meta-analysis of the effectiveness of albendazole compared with metronidazole as treatments for infections with Giardia duodenalis. PLoS Negl Trop Dis. 2010;4(5):e682.
10. Goldman P, Koch RL, Yeung TC, et al. Comparing the reduction of nitroimidazoles in bacteria and mammalian tissues and relating it to biological activity. Biochem Pharmacol. 1986;35(1):43–51.
11. Tian HF, Chen B, Wen JF. Giardiasis, drug resistance, and new target discovery. Infect Disord Drug Targets. 2010;10(4):295–302.
12. Leitsch D, Schlosser S, Burgess A, Duchene M. Nitroimidazole drugs vary in their mode of action in the human parasite Giardia lamblia. Int J Parasitol Drugs Drug Resist. 2012;2:166–170. doi:10.1016/j.ijpddr.2012.04.002
13. Uzlikova M, Nohynkova E. The effect of metronidazole on the cell cycle and DNA in metronidazole-susceptible and -resistant Giardia cell lines. Mol Biochem Parasitol. 2014;198(2):75–81. doi:10.1016/j.molbiopara.2015.01.005
14. Upcroft P, Upcroft JA. Drug targets and mechanisms of resistance in the anaerobic protozoa. Clin Microbiol Rev. 2001;14(1):150–164. doi:10.1128/CMR.14.1.150-164.2001
15. Raj D, Ghosh E, Mukherjee AK, Nozaki T, Ganguly S. Differential gene expression in Giardia lamblia under oxidative stress: significance in eukaryotic evolution. Gene. 2014;535(2):131–139. doi:10.1016/j.gene.2013.11.048
16. Ansell BR, Baker L, Emery SJ, et al. Transcriptomics indicates active and passive metronidazole resistance mechanisms in three seminal Giardia lines. Front Microbiol. 2017;8:398. doi:10.3389/fmicb.2017.00398
17. Muller J, Rout S, Leitsch D, Vaithilingam J, Hehl A, Muller N. Comparative characterisation of two nitroreductases from Giardia lamblia as potential activators of nitro compounds. Int J Parasitol Drugs Drug Resist. 2015;5(2):37–43. doi:10.1016/j.ijpddr.2015.03.001
18. Muller J, Schildknecht P, Muller N. Metabolism of nitro drugs metronidazole and nitazoxanide in Giardia lamblia: characterization of a novel nitroreductase (GlNR2). J Antimicrob Chemother. 2013;68(8):1781–1789. doi:10.1093/jac/dkt106
19. Nillius D, Muller J, Muller N. Nitroreductase (GlNR1) increases susceptibility of Giardia lamblia and Escherichia coli to nitro drugs. J Antimicrob Chemother. 2011;66(5):1029–1035. doi:10.1093/jac/dkr029
20. Mastronicola D, Falabella M, Forte E, Testa F, Sarti P, Giuffre A. Antioxidant defence systems in the protozoan pathogen Giardia intestinalis. Mol Biochem Parasitol. 2016;206(1–2):56–66. doi:10.1016/j.molbiopara.2015.12.002
21. Holmes AH, Moore LS, Sundsfjord A, et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet (London, England). 2016;387(10014):176–187. doi:10.1016/S0140-6736(15)00473-0
22. Munoz Gutierrez J, Aldasoro E, Requena A, et al. Refractory giardiasis in Spanish travellers. Travel Med Infect Dis. 2013;11(2):126–129. doi:10.1016/j.tmaid.2012.10.004
23. Requena-Mendez A, Goni P, Rubio E, et al. The use of quinacrine in nitroimidazole-resistant Giardia duodenalis: an old drug for an emerging problem. J Infect Dis. 2017;215(6):946–953. doi:10.1093/infdis/jix066
24. Nabarro LE, Lever RA, Armstrong M, Chiodini PL. Increased incidence of nitroimidazole-refractory giardiasis at the Hospital for Tropical Diseases, London: 2008–2013. Clin Microbiol Infect. 2015;21(8):791–796. doi:10.1016/j.cmi.2015.04.019
25. Meltzer E, Lachish T, Schwartz E. Treatment of giardiasis after nonresponse to nitroimidazole. Emerg Infect Dis. 2014;20(10):1742–1744. doi:10.3201/eid2010.140073
26. Requena-Mendez A, Goni P, Lobez S, et al. A family cluster of giardiasis with variable treatment responses: refractory giardiasis in a family after a trip to India. Clin Microbiol Infect. 2014;20(2):O135–O138. doi:10.1111/1469-0691.12327
27. Cruz A, Sousa MI, Azeredo Z, Leite E, Figueiredo de Sousa JC, Cabral M. Isolation, excystation and axenization of Giardia lamblia isolates: in vitro susceptibility to metronidazole and albendazole. J Antimicrob Chemother. 2003;51(4):1017–1020. doi:10.1093/jac/dkg150
28. Mastronicola D, Giuffre A, Testa F, et al. Giardia intestinalis escapes oxidative stress by colonizing the small intestine: a molecular hypothesis. IUBMB Life. 2011;63(1):21–25. doi:10.1002/iub.409
29. Lebbad M, Petersson I, Karlsson L, et al. Multilocus genotyping of human Giardia isolates suggests limited zoonotic transmission and association between assemblage B and flatulence in children. PLoS Negl Trop Dis. 2011;5(8):e1262. doi:10.1371/journal.pntd.0001370
30. Townson SM, Upcroft JA, Upcroft P. Characterisation and purification of pyruvate: ferredoxinoxidoreductase from Giardia duodenalis. Mol Biochem Parasitol. 1996;79(2):183–193.
31. Tejman-Yarden N, Millman M, Lauwaet T, et al. Impaired parasite attachment as fitness cost of metronidazole resistance in Giardia lamblia. Antimicrob Agents Chemother. 2011;55(10):4643–4651. doi:10.1128/AAC.00384-11
32. Leitsch D, Burgess AG, Dunn LA, et al. Pyruvate:ferredoxinoxidoreductase and thioredoxin reductase are involved in 5-nitroimidazole activation while flavin metabolism is linked to 5-nitroimidazole resistance in Giardia lamblia. J Antimicrob Chemother. 2011;66(8):1756–1765. doi:10.1093/jac/dkr192
33. Dan M, Wang AL, Wang CC. Inhibition of pyruvate-ferredoxin oxidoreductase gene expression in Giardia lamblia by a virus-mediated hammerhead ribozyme. Mol Microbiol. 2000;36(2):447–456.
34. Muller J, Wastling J, Sanderson S, Muller N, Hemphill A. A novel Giardia lamblia nitroreductase, GlNR1, interacts with nitazoxanide and other thiazolides. Antimicrob Agents Chemother. 2007;51(6):1979–1986. doi:10.1128/AAC.01548-06
35. Emery SJ, Baker L, Ansell BRE, et al. Differential protein expression and post-translational modifications in metronidazole-resistant Giardia duodenalis. GigaScience. 2018;7:4. doi:10.1093/gigascience/giy024
36. Ansell BR, McConville MJ, Ma’ayeh SY, et al. Drug resistance in Giardia duodenalis. Biotechnol Adv. 2015;33(6 Pt 1):888–901. doi:10.1016/j.biotechadv.2015.04.009
37. Ma’ayeh SY, Knorr L, Svard SG. Transcriptional profiling of Giardia intestinalis in response to oxidative stress. Int J Parasitol. 2015;45(14):925–938. doi:10.1016/j.ijpara.2015.07.005
38. Leitsch D. A review on metronidazole: an old warhorse in antimicrobial chemotherapy. Parasitology. 2017;1–12. doi:10.1017/S0031182017002025
39. Leitsch D. Drug resistance in the microaerophilic parasite Giardia lamblia. Curr Trop Med Rep. 2015;2(3):128–135. doi:10.1007/s40475-015-0051-1
40. Begaydarova R, Yukhnevich Y, Babenko D, et al. Determination of PFOR gene expression in strains of G. intestinalis with different inhibitory concentrations of metronidazole. J Infect Dev Ctries. 2015;9(5):519–523. doi:10.3855/jidc.5768
41. Galeh TM, Kazemi A, Mahami-Oskouei M, et al. Introducing nitazoxanide as a promising alternative treatment for symptomatic to metronidazole-resistant giardiasis in clinical isolates. Asian Pac J Trop Med. 2016;9(9):887–892. doi:10.1016/j.apjtm.2016.07.013
42. Gabin-Garcia LB, Bartolome C, Abal-Fabeiro JL, Mendez S, Llovo J, Maside X. Strong genetic structure revealed by multilocus patterns of variation in Giardia duodenalis isolates of patients from Galicia (NW-Iberian Peninsula). Infect. Genet. Evol. 2016;48:131–141. doi:10.3855/jidc.5768
43. Teodorovic S, Braverman JM, Elmendorf HG. Unusually low levels of genetic variation among Giardia lamblia isolates. Eukaryot Cell. 2007;6(8):1421–1430. doi:10.1128/EC.00138-07
44. Muller J, Ley S, Felger I, Hemphill A, Muller N. Identification of differentially expressed genes in a Giardia lamblia WB C6 clone resistant to nitazoxanide and metronidazole. J Antimicrob Chemother. 2008;62(1):72–82. doi:10.1093/jac/dkn142
45. Hanevik K, Bakken R, Brattbakk HR, Saghaug CS, Langeland N. Whole genome sequencing of clinical isolates of Giardia lamblia. Clin Microbiol Infect. 2015;21(2):
46. Adam RD, Dahlstrom EW, Martens CA, et al. Genome sequencing of Giardia lamblia genotypes A2 and B isolates (DH and GS) and comparative analysis with the genomes of genotypes A1 and E (WB and Pig). Genome Biol Evol. 2013;5(12):2498–2511. doi:10.1093/gbe/evt197
47. Ansell BR, McConville MJ, Baker L, et al. Divergent transcriptional responses to physiological and xenobiotic stress in Giardia duodenalis. Antimicrob Agents Chemother. 2016;60(10):6034–6045. doi:10.1128/AAC.00977-16
48. Nixon JE, Wang A, Field J, et al. Evidence for lateral transfer of genes encoding ferredoxins, nitroreductases, NADH oxidase, and alcohol dehydrogenase 3 from anaerobic prokaryotes to Giardia lamblia and Entamoeba histolytica. Eukaryot Cell. 2002;1(2):181–190.
49. Rice EW, Schaefer FW
50. Keister DB. Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans R Soc Trop Med Hyg. 1983;77(4):487–488.
51. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi:10.1038/nmeth.1923
52. Aurrecoechea C, Brestelli J, Brunk BP, et al. GiardiaDB and TrichDB: integrated genomic resources for the eukaryotic protist pathogens Giardia lamblia and Trichomonas vaginalis. Nucleic Acids Res. 2009;37(Database issue):D526–D530. doi:10.1093/nar/gkn631
53. Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–858. doi:10.1038/nprot.2015.053
54. Nixon JE, Wang A, Morrison HG, et al. A spliceosomal intron in Giardia lamblia. Proc Natl Acad Sci U S A. 2002;99(6):3701–3705.
55. Fukuyama K, Matsubara H, Tsukihara T, Katsube Y. Structure of [4Fe-4S] ferredoxin from Bacillus thermoproteolyticus refined at 2.3 A resolution. Structural comparisons of bacterial ferredoxins. J Mol Biol. 1989;210(2):383–398.
56. Duee ED, Fanchon E, Vicat J, Sieker LC, Meyer J, Moulis JM. Refined crystal structure of the 2[4Fe-4S] ferredoxin from Clostridium acidurici at 1.84 A resolution. J Mol Biol. 1994;243(4):683–695.
57. Franzen O, Jerlstrom-Hultqvist J, Castro E, et al. Draft genome sequencing of giardia intestinalis assemblage B isolate GS: is human giardiasis caused by two different species? PLoS Pathog. 2009;5(8):e1000560.
58. Thompson A. Human giardiasis: genotype-linked differences in clinical symptomatology. Trends Parasitol. 2001;17(10):465.
59. Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. 2011;24(1):110–140.
60. Ellis JE, Wingfield JM, Cole D, Boreham PF, Lloyd D. Oxygen affinities of metronidazole-resistant and -sensitive stocks of Giardia intestinalis. Int J Parasitol. 1993;23(1):35–39.
61. Smith NC, Bryant C, Boreham PF. Possible roles for pyruvate: ferredoxinoxidoreductase and thiol-dependent peroxidase and reductase activities in resistance to nitroheterocyclic drugs in Giardia intestinalis. Int J Parasitol. 1988;18(7):991–997.
62. Goodwin A, Kersulyte D, Sisson G, Veldhuyzen van Zanten SJ, Berg DE, Hoffman PS. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol. 1998;28(2):383–393.
63. Sisson G, Jeong JY, Goodwin A, et al. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and in Escherichia coli containing a cloned H. pylori RdxA(+) (Nitroreductase) gene. J Bacteriol. 2000;182(18):5091–5096.
64. Kobori T, Sasaki H, Lee WC, et al. Structure and site-directed mutagenesis of a flavoprotein from Escherichia coli that reduces nitrocompounds: alteration of pyridine nucleotide binding by a single amino acid substitution. J Biol Chem. 2001;276(4):2816–2823.
65. Paulish-Miller TE, Augostini P, Schuyler JA, et al. Trichomonas vaginalis metronidazole resistance is associated with single nucleotide polymorphisms in the nitroreductase genes ntr4Tv and ntr6Tv. Antimicrob Agents Chemother. 2014;58(5):2938–2943.
66. Bradic M, Warring SD, Tooley GE, et al. Genetic indicators of drug resistance in the highly repetitive genome of Trichomonas vaginalis. Genome Biol Evol. 2017;9(6):1658–1672.
67. Testa F, Mastronicola D, Cabelli DE, et al. The superoxide reductase from the early diverging eukaryote Giardia intestinalis. Free Radic Biol Med. 2011;51(8):1567–1574.
68. Kwon DH, El-Zaatari FA, Kato M, et al. Analysis of rdxA and involvement of additional genes encoding NAD(P)H flavin oxidoreductase (FrxA) and ferredoxin-like protein (FdxB) in metronidazole resistance of Helicobacter pylori. Antimicrob Agents Chemother. 2000;44(8):2133–2142.
69. Caccio SM, Ryan U. Molecular epidemiology of giardiasis. Mol Biochem Parasitol. 2008;160(2):75–80.
70. Marais A, Bilardi C, Cantet F, Mendz GL, Megraud F. Characterization of the genes rdxA and frxA involved in metronidazole resistance in Helicobacter pylori. Res Microbiol. 2003;154(2):137–144.
71. Mora A, Sandve GK, Gabrielsen OS, Eskeland R. In the loop: promoter-enhancer interactions and bioinformatics. Brief Bioinform. 2016;17(6):980–995.
72. Elmendorf HG, Singer SM, Pierce J, Cowan J, Nash TE. Initiator and upstream elements in the alpha2-tubulin promoter of Giardia lamblia. Mol Biochem Parasitol. 2001;113(1):157–169.
73. Mastronicola D, Testa F, Forte E, et al. Flavohemoglobin and nitric oxide detoxification in the human protozoan parasite Giardia intestinalis. Biochem Biophys Res Commun. 2010;399(4):654–658.
74. Gardner PR, Gardner AM, Martin LA, Salzman AL. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc Natl Acad Sci U S A. 1998;95(18):10378–10383.
75. Hausladen A, Gow AJ, Stamler JS. Nitrosative stress: metabolic pathway involving the flavohemoglobin. Proc Natl Acad Sci U S A. 1998;95(24):14100–14105.
76. Mastronicola D, Falabella M, Testa F, et al. Functional characterization of peroxiredoxins from the human protozoan parasite Giardia intestinalis. PLoS Negl Trop Dis. 2014;8(1):e2631.
77. Martinez-Julvez M, Rojas AL, Olekhnovich I, Espinosa Angarica V, Hoffman PS, Sancho J. Structure of RdxA–an oxygen-insensitive nitroreductase essential for metronidazole activation in Helicobacter pylori. FEBS J. 2012;279(23):4306–4317.
78. Townson SM, Hanson GR, Upcroft JA, Upcroft P. A purified ferredoxin from Giardia duodenalis. Eur J Biochem/FEBS. 1994;220(2):439–446.
79. Di Matteo A, Scandurra FM, Testa F, et al. The O2-scavenging flavodiiron protein in the human parasite Giardia intestinalis. J Biol Chem. 2008;283(7):4061–4068.
80. Leitsch D, Muller J, Muller N. Evaluation of Giardia lamblia thioredoxin reductase as drug activating enzyme and as drug target. Int J Parasitol Drugs Drug Resist. 2016;6(3):148–153.
81. Brown DM, Upcroft JA, Edwards MR, Upcroft P. Anaerobic bacterial metabolism in the ancient eukaryote Giardia duodenalis. Int J Parasitol. 1998;28(1):149–164.
82. Hofstetrova K, Uzlikova M, Tumova P, Troell K, Svard SG, Nohynkova E. Giardia intestinalis: aphidicolin influence on the trophozoite cell cycle. Exp Parasitol. 2010;124(2):159–166.
83. Sanchez LB. Aldehyde dehydrogenase (CoA-acetylating) and the mechanism of ethanol formation in the amitochondriate protist, Giardia lamblia. Arch Biochem Biophys. 1998;354(1):57–64.
84. Brown DM, Upcroft JA, Upcroft P. A H2O-producing NADH oxidase from the protozoan parasite Giardia duodenalis. Eur J Biochem/FEBS. 1996;241(1):155–161.
85. Sanchez LB, Elmendorf H, Nash TE, Muller M. NAD(P)H:menadioneoxidoreductase of the amitochondriate eukaryote Giardia lamblia: a simpler homologue of the vertebrate enzyme. Microbiology. 2001;147(Pt 3):561–570.
86. Muller J, Sterk M, Hemphill A, Muller N. Characterization of Giardia lamblia WB C6 clones resistant to nitazoxanide and to metronidazole. J Antimicrob Chemother. 2007;60(2):280–287.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.