Back to Journals » Cancer Management and Research » Volume 13
Genetic Variants in METTL14 are Associated with the Risk of Acute Lymphoblastic Leukemia in Southern Chinese Children: A Five-Center Case-Control Study
Authors Luo A, Yang L, Li M, Cai M, Huang A, Liu X, Yang X, Yan Y, Wang X, Wu X, Huang K, Huang L, Liu S, Xu L, Liu X
Received 26 August 2021
Accepted for publication 12 November 2021
Published 14 December 2021 Volume 2021:13 Pages 9189—9200
DOI https://doi.org/10.2147/CMAR.S335925
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Bilikere Dwarakanath
Ailing Luo,1,* Lihua Yang,2,* Ming Li,3 Mansi Cai,1 Amin Huang,4 Xiaodan Liu,5 Xu Yang,1 Yaping Yan,1 Xueliang Wang,1 Xuedong Wu,6 Ke Huang,7 Libin Huang,8 Shanshan Liu,1 Ling Xu,1 Xiaoping Liu1
1Department of Hematology, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou, Guangdong, People’s Republic of China; 2Pediatric Center of Zhujiang Hospital, Southern Medical University, Guangzhou, Guangdong, People’s Republic of China; 3The Emergency Department of Zhuzhou Central Hospital, Zhuzhou, Hunan, People’s Republic of China; 4Department of Medical Oncology, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, People’s Republic of China; 5Division of Birth Cohort Study, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou, Guangdong, People’s Republic of China; 6Department of Pediatrics, Nanfang Hospital, Southern Medical University, Guangzhou, Guangdong, People’s Republic of China; 7Department of Pediatrics, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, Guangdong, People’s Republic of China; 8Pediatrics Department, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaoping Liu; Ling Xu
Department of Hematology, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, No. 9, Jinsui Road, Tianhe District, Guangzhou, 510623, Guangdong, People’s Republic of China
Tel +86-13822283646
; +86-13798111250
Fax +86-2038076069
; +86-2038076557
Email [email protected]; [email protected]
Background/Aim: Acute lymphoblastic leukemia (ALL) is the most common form of pediatric cancer. METTL14, an N6-methyladenosine (m6A) modification protein, plays several roles in cancer development and is involved in the pathogenesis of various types of cancers. However, the role of METTL14 gene single nucleotide polymorphisms (SNPs) in pediatric ALL susceptibility remains to be investigated.
Methods: A case-control design and multinomial logistic regression were used to develop models to estimate the overall risk for pediatric ALL and three METTL14 gene SNPs (rs298982 G/A, rs298981 A/G and rs1064034 T/A) in 808 cases and 1340 controls, which were genotyped using a TaqMan assay. The associations were estimated by odds ratios (ORs) with their 95% confidence intervals (CIs). Furthermore, stratified analysis was performed to explore associations of rs298982 and rs1064034 with pediatric ALL susceptibility in terms of age, sex, immunophenotype, minimal residual disease (MRD), and other clinical characteristics.
Results: Among the three analyzed SNPs, rs298982 G/A and rs1064034 T/A exhibited a significant association with decreased childhood ALL risk, while rs298981 A/G exhibited no difference. In stratified analysis, rs298982 GA/AA and rs1064034 TA/AA had a protective effect in children < 120 months of age and males, common B ALL, TEL-AML, non gene fusion, normal diploid, and high WBC. However, the rs1064034 TA/AA genotype was associated with an increased risk of mixed immunophenotyping. Compared with the reference haplotype GAT, haplotypes CAA, CGT and CGA were significantly associated with elevated ALL risk, while haplotype GGT was significantly associated decreased ALL risk. Moreover, subjects carrying rs298982 A or rs1064034 A exhibited less minimal MRD after induced chemotherapy. Functional annotations revealed that METTL14 gene SNPs rs298982 G/A and rs1064034 T/A could be potential functional variants.
Conclusion: In conclusion, METTL14 gene polymorphisms influence the risk of ALL in southern Chinese children and might be potential biomarkers for pediatric ALL susceptibility and chemotherapeutics.
Keywords: ALL, METTL14, SNP, susceptibility, Chinese children
Introduction
Acute lymphoblastic leukemia (ALL) is the most common form of pediatric cancer, accounting for approximately 25% of childhood cancers and nearly 80% of all leukemia cases.1,2 ALL primarily leads to changes in the lymphocyte composition in the blood, resulting in the replacement of mature functional lymphocytes with immature lymphocytes.3 ALL arises from hematopoietic cells in either the B-cell precursor (B-ALL) or T-cell lineages (T-ALL).4,5 Bone marrow transplantation and targeted drugs are preferred for ALL treatment, but the economic and human resource cost is enormous.3 Investigating the pathogenesis of ALL may facilitate the complete cure of the disease to some extent. Genetic changes play a key role in the childhood prevalence of ALL.6 Gene polymorphisms were found to be closely associated with ALL sensitivity. Mosaad et al found that ARID5B rs10821936 and the CA haplotype can be considered susceptibility risk factors for the development of pediatric ALL and adult ALL in the studied cohort of Egyptian patients.1 Yang et al revealed that FOXO3 gene polymorphisms influence the risk of ALL in children and might be a potential biomarker for ALL susceptibility.7
N6-methyladenosine (m6A) is the most abundant internal modification in mRNAs and lncRNAs in eukaryotic cells. The m6A modification process is accomplished by a series of proteins, which include methyltransferases (writers), demethylases (erasers) and binding proteins (readers). M6A modification has been reported to play multiple roles in cancer development and to be involved in the pathogenesis of various types of cancers.8–10 However, there were few studies about the role of METTL14 in leukemia. METTL14, an m6A “writer”, was found a high expression level in normal hematopoietic stem/progenitor cells (HSPCs) and in AMLs, when its depletion could promote differentiation of HSPCs and AML cells.11 And to date, only one studies have characterized METTL14 in childhood patients with ALL. Sun et al found that the expression levels of METTL3 and METTL14 in E/R-positive ALL patients were lower than those in controls, which suggested that METTL3 and METTL14 expression may be a new prognostic factor and indicate specific treatment intensification in possible E/R-positive relapse patients.12 Previously, we reported that multiple m6A-modified enzyme-encoding genes, METTL3, METTL14, WTAP, FTO, and ALKBH5, are closely related to glioma susceptibility. METTL14 polymorphisms have been reported to increase susceptibility to neuroblastoma.13 Based on these findings, we hypothesized that polymorphisms of METTL14 gene are associated with the risk of pediatric ALL.
To identify novel ALL susceptibility single nucleotide polymorphisms (SNPs) of METTL14 gene, an m6A modification key regulator, we conducted a five-center hospital-based case-control study. We addressed the associations between three METTL14 gene SNPs and ALL risk in a population of Chinese children.
Materials and Methods
Patients and Healthy Controls
In this study, participants were divided into 2 groups: childhood ALL patients and healthy controls. A total of 808 patients and 1340 controls, with an age range of 0.5 to 17 years old, were recruited from Guangzhou Women and Children’s Center, Sun Yat-sen Memory Hospital Affiliated Sun Yat-sen University (SYSU), Nanfang Hospital Affiliated Southern Medical University, The First Hospital Affiliated SYSU and Zhujiang Hospital Affiliated Southern Medical University. There were no statistically significant differences in age, sex, or other general clinical data between the ALL group and the control group (P>0.05) as described previously.14 Informed consent was obtained from the parents of children with ALL. All patients were diagnosed according to standard methods, including cytomorphologic, cytochemical, and immunophenotyping methods. ALL patients without previous therapy were included in the study. Control subjects, who were recruited concurrently with ALL patient subjects, were randomly selected from the volunteers visiting the hospital and matched according to the expected age and gender distribution of patients. The markers of CD38/CD81, CD66c/CD34/CD10/CD19/CD45 were used for B-ALL; and markers of CD3/CD99/CD19, CD33/HLA-DR/CD5/CD7/CD45, CD10/CD19, CD33/HLA-DR/CD5/CD7/CD45 were used to for T-ALL to detect MRD by flow cytometry. The institutional review board of Guangzhou Women and Children’s Medical Center gave approval for the current study.
SNP Selection and Genotyping
DNA was extracted from EDTA blood using the TIANamp DNA Kit (Tiangen, Beijing, China) according to the standard instructions of the kit. We used NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP) and SNPinfo (http://snpinfo.niehs.nih.gov/snpfunc.htm) to select the candidate SNPs of METTL14 which is potentially functional, which follows the previously described criteria: located in the 5ʹ untranslated region, 3ʹ untranslated region, 5ʹ flanking region, and exon of METTL14 gene.15 Three METTL14 gene SNPs, rs298982 G/A, rs298981 A/G and rs1064034 T/A, were chosen. TaqMan genotyping kits (Tiangen, Beijing, China) were used for SNP genotyping on an ABI 7900 (Applied Biosystem, Foster City, CA, USA) as described previously.16–18 Genotyping results were confirmed by randomly assaying 10% of the original specimens for replication to exclude genotyping errors. There were no discrepancies between genotypes determined in duplicate. Further functional annotation of the significant SNPs was performed using Genotype-Tissue Expression (GTEx) (https://gtexportal.org). All tests for statistical significance used a two-sided alpha of 0.05. All statistical analyses were conducted using SAS v10.0 (SAS Institute, Cary, NC, USA).
Statistical Analyses
The compliance of genotypes with HWE among controls was appraised by a χ2 test. Differences in demographic characteristics between the cases and the controls were evaluated by the χ2 test. The age- and sex-adjusted ORs and 95% CIs for the relationships between the SNPs and ALL risk were determined by multivariate logistic regression analysis. Logistic regression analyses were adopted to estimation haplotype frequencies and their effect on ALL risk, with the adjustment for gender and age.19,20 To determine whether the significant findings were “noteworthy,” false-positive report probability (FPRP) analysis was performed for each significant finding as described previously.21 A prior probability of 0.1 was adopted to detect odds ratios (ORs) of 1.50 and 0.67 for risk and protective effects, respectively. An association with an FPRP value < 0.2 was considered noteworthy. All statistical analyses were conducted using SAS v10.0 (SAS Institute, Cary, NC, USA). In the present study, all P values were two-sided, and values of P<0.05 were considered statistically significant.
Results
Association of METTL14 Gene SNPs on Pediatric ALL Risk
Three METTL14 gene SNPs (rs298982 G/A, rs298981 A/G and rs1064034 T/A) were successfully genotyped in 808 cases and 1340 controls. Table 1 shows the association of these three SNPs with pediatric ALL risk. All SNPs were in HWE in the two groups (P>0.05). Of the three SNPs, in a single–locus analysis, carriers of the rs298982 (GA versus GG: adjusted odds ratio [OR]=0.66, 95% confidence interval [CI]=0.52–0.84, P=0.001; AA/GA versus GG: adjusted OR=0.69, 95% CI=0.55–0.87, P=0.002) and rs1064034 (TA versus TT: adjusted OR=0.70, 95% CI=0.58–0.86, P=0.001; AA/TA versus TT: adjusted OR=0.72, 95% CI=0.60–0.87, P=0.001) variant alleles showed decreased susceptibility to ALL. However, the remaining genotype, rs298981 A/G, was not associated with ALL risk. We then defined rs298982 GG/GA and rs1064034 TT/TA as risk genotypes.
 |
Table 1 Logistic Regression Analysis of Associations Between METTL14 Polymorphisms and ALL Susceptibility |
Stratification Analysis of rs298982 and rs1064034 with ALL Susceptibility
Subgroup analyses by age, sex, immunophenotyping, gene infusion, karyotype, WBC, MRD and other clinical information to evaluate the effect of rs298982 G/A and rs1064034 T/A is shown in Table 2. We found that rs298982 GA/AA had a protective effect in children <120 months of age (adjusted OR=0.70, 95% CI=0.55–0.90, P=0.004) and males (adjusted OR=0.69, 95% CI=0.51–0.92, P=0.013). We also found that rs298982 GA/AA decreased the ALL risk in patients with common B ALL (adjusted OR=0.36, 95% CI=0.24–0.56, P=0.001), TEL-AML (adjusted OR=0.56, 95% CI=0.33–0.93, P=0.025), non gene fusion (adjusted OR=0.75, 95% CI=0.58–0.98, P=0.031), normal diploid (adjusted OR=0.67, 95% CI=0.51–0.89, P=0.005), primitive/naïve lymphocytes in marrow <5% (adjusted OR=0.71, 95% CI=0.54–0.93, P=0.013), high WBC (adjusted OR=0.59, 95% CI=0.41–0.83, P=0.003) and with MRD ≥0.01 on Day 33 after induced therapy (adjusted OR=0.40, 95% CI=0.25–0.64, P=0.001).
 |
Table 2 Stratification Analysis of METTL14 Polymorphisms with ALL Susceptibility |
Rs1064034 TA/AA had a protective effect in children <120 months of age (adjusted OR=0.74, 95% CI=0.61–0.90, P=0.003) and males (adjusted OR=0.71; 95% CI=0.56–0.90, P=0.004). Compared to the TT genotype, the rs1064034 TA/AA genotype was associated with a decreased risk of common B ALL (adjusted OR=0.58, 95% CI=0.44–0.78, P=0.001), pre B ALL (adjusted OR=0.71; 95% CI=0.50–0.99, P=0.046), T ALL (adjusted OR=0.54; 95% CI=0.30–0.95, P=0.032), MLL (adjusted OR=0.26; 95% CI=0.07–0.94, P=0.040), non gene fusion type (adjusted OR=0.74; 95% CI=0.60–0.91, P=0.005), high WBC (adjusted OR=0.64, 95% CI=0.49–0.84, P=0.002) and normal diploid (adjusted OR=0.76; 95% CI=0.61–0.94, P=0.011). In addition, rs1064034 T/A decreased ALL risk in patients with primitive/naïve lymphocytes in marrow <5% on Day 33 (adjusted OR=0.67; 95% CI=0.54–0.84, P=0.001) and Week 12 (adjusted OR=0.77; 95% CI=0.60–0.99, P=0.040), MRD ≥0.01 on Day 33 (adjusted OR=0.42; 95% CI=0.30–0.60, P=0.001) and on Week 12 (adjusted OR=0.05; 95% CI=0.007–0.39, P=0.004). However, the rs1064034 TA/AA genotype was associated with an increased risk of mixed immunophenotyping (adjusted OR=1.88; 95% CI=1.08–3.27, P=0.026).
Haplotype Analysis of Three METTL14 Gene SNPs Correlated with ALL Susceptibility
We further determined whether the haplotypes of the three METTL14 gene SNPs are linked to ALL risk. As shown in Table 3, the wildtype allele GAT was defined as the reference group. Compared with the reference haplotype GAT, the following haplotypes were significantly associated with elevated ALL risk: CAA (adjusted OR=5.27, 95% CI=4.25–6.53, P<0.001), CGT (adjusted OR=1.64, 95% CI=1.24–2.17, P=0.001) and CGA (adjusted OR=6.83, 95% CI=5.59–8.34, p<0.001). While haplotype GGT (adjusted OR=0.12, 95% CI=0.08–0.16, P<0.001) was significantly associated decreased ALL risk.
 |
Table 3 Association Between Inferred Haplotypes of the METTL14 Gene and ALL Risk |
The Influence of Identified SNPs on Sensitivity to Different Treatment Strategies Based on MRD Levels
We further assessed the MRD in the marrow of patients with different alleles after treatment with Chinese Children Cancer Group chemotherapeutics (CCCGs) or South China Children Leukemia Group chemotherapeutics (SCCLGs) (Tables 4 and 5). For rs298982, GG alleles had a harmful effect on MRD in marrow ≥0.01% at 33 d (adjusted OR=3.00, 95% CI=1.66–5.44, P=0.001) after CCCG treatment. For rs1064034, TT alleles had a harmful effect on MRD in marrow ≥0.01% at 33 d (adjusted OR=2.32, 95% CI=1.45–3.69, P=0.001) after CCCG treatment. These results indicated that southern Chinese pediatric patients with rs298982 GA/AA alleles or rs1064034 TA/AA alleles may have better outcomes after receiving CCCG treatment.
 |
Table 4 The Influence of METTL14 Polymorphisms (rs298982, GA/AA vs GG) on Sensitivity to Different Treatment Strategies Based on MRD Levels |
 |
Table 5 The Influence of METTL14 Polymorphisms (rs1064034, TA/AA vs TT) on Sensitivity to Different Treatment Strategies Based on MRD Levels |
Expression Quantitative Trait Loci (eQTL) Analyses
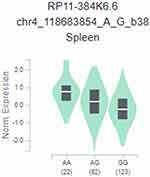
We further assessed the putative functional relevance of METTL14 rs298982 and rs1064034 using released data from GTEx. Analysis revealed that the rs298982 A genotype was associated with significantly increased RP11-3846.6 expression level in spleen (Figure 1). In addition to the METTL14 rs298982 A genotype, samples with the rs1064034 T genotype had a higher lncSNHG8 (long noncoding RNA SNHG8) expression level in the cultured fibroblasts than the A allele (Figure 2A). Our cis-eQTL analysis also detected an association between the rs1064034 A genotype and increased expression of the RP11-3846.6 gene in whole blood and cultured fibroblasts (Figure 2B and C).
Discussion
In the present case-control study, we investigated the potential association of METTL14 polymorphisms with ALL risk between 808 ALL patients and 1340 healthy controls from a population of southern Chinese children. We showed that two of the three selected SNPs were associated with ALL risk: both rs298982 G/A and rs1064034 T/A were related to decreased ALL risk. To the best of our knowledge, this is the first study on the association between the METTL14 rs298982 and rs1064034 polymorphisms and ALL risk.
METTL14, methyltransferase like 14, a methyltransferase, has been reported to act as an oncogene by affecting RNA stability and degradation through pre-RNA splicing, protein translation, and miRNA processing in a variety of tumors.11,22,23 Sun et al reported that METTL14 was significantly upregulated in breast cancer and that a novel LNC942-METTL14-CXCR4/CYP1B1 signaling axis was involved in BRCA prevention and treatment.24 Chen et al detected a lower expression level of METTL14 in colorectal cancer tissues and cell lines by modulating the N6-methyladenosine level of SOX4 mRNA, which was significantly associated with poor overall survival.23 Yi et al identified the potential effect of METTL14 on the miRNA expression profile and the effect of hsa-miR-146a-5p on the migration and invasion of breast cancer cells. Weng et al found that METTL14 was highly expressed in acute myeloid leukemia cells and played an oncogenic role.11 However, there are few studies about the association of METTL14 genetic variants with cancer risk. Zhou et al investigated four METTL14 gene SNPs (rs298982 G/A, rs62328061 A/G, rs9884978 G/A, and rs4834698 T/C) associated with neuroblastoma susceptibility.13 In our previous research, we reported that multiple m6A-modified enzyme-encoding genes are closely related to glioma susceptibility, and none of the four studied METTL14 gene SNPs (rs1064034, rs298981, rs62328061, and rs9884978) were associated with glioma risk in Chinese children.15
In the present work, we performed genotyping of three SNPs of METTL14, rs298982, rs298981 and rs1064034 and demonstrated that rs298982 G/A and rs1064034 T/A were associated with pediatric ALL risk. Our results suggested that the rs298982 A allele and rs1064034 A allele decreased the ALL risk in a population of southern Chinese children. Both rs298982 and rs1064034 play a role in decreased ALL risk predominantly in children < 10 years and males and in patients with common B immunophenotyping, normal diploid or positive MRD in CCCGs, rather in SCCLG. Moreover, the effects of rs1064034 on ALL risk were also predominant in patients with T-ALL and MLL fusion types. These results suggested that rs298982 G/A and rs1064034 T/A may provide a reference for the diagnosis and therapy of this disease. Compared with single SNPs, multiple markers association studies based on haplotypes significantly improve the power of mapping and characterizing disease-causing genes.25 We explored whether various haplotypes consisting of the METTL14 gene polymorphisms rs298982, rs298981 and rs1064034 are associated with ALL risk. These results suggest that these variants may interact with each other to modify the risk of ALL. We further attempted to interpret the possible mechanism of METTL14 gene SNP-mediated ALL risk. Furthermore eQTL evidence suggested that the A allele in rs298982 are associated with increased RP11-384K6.6 levels in spleen. We also revealed that the higher expression of lncSNHG8 caused by the T allele in rs1064034 in cultured fibroblasts but decreased expression of RP11-384K6.6 in whole blood and c cultured fibroblasts. LncSNHG8 was found to be abnormally expressed in cancers and to play important roles in several kinds of cancers.26–28 Tian et al found LncSNHG8 higher expression level in nasopharyngeal carcinoma tissues and cells, as well as it regulates nasopharyngeal carcinoma cell proliferation, colony formation, migration, and invasion via regulating miR-656-3p.27 Miao et al demonstrated that lncSNHG8 expression was elevated in ovarian carcinoma cells, and revealed that silencing of it impedes cell proliferation and promote cell apoptosis.29 Furthermore, Dong et al found that lncSNHG8 expression was increased in human hepatocellular carcinoma tissues which promoted cell migration and invasion.30 But there are no research about LncSNHG8 in ALL and RP11-384K6.6 in cancers, We propose that the higher expression of lncSNHG8 caused by the A allele in rs298982 and the T allele in rs1064034 may facilitate the development of ALL. On the other hand, with increases in the rs1064034 A genotype, the average expression of RP11-384K6.6 gradually increased. This conclusion requires further interpretation because the role of RP11-384K6.6 in cancers remains to be revealed. In conclusion, more functional experiments are needed to support this possible mechanism.
Although this study explored METTL14 polymorphisms and ALL susceptibility in a novel and relatively large sample size of pediatric acute lymphoblastic leukemia cases and included multiple-center participants in a single Chinese population, several limitations should be noted. First, the study population involved only Chinese subjects and was limited to volunteers. Larger-scale studies with more samples and SNP sites are encouraged to confirm the roles of METTL14 in ALL risk. Finally, this study only analyzed genetic factors, and other environmental factors were not available for ALL susceptibility investigation. The functions of METTL14 and these SNPs in the progression of ALL should also be further investigated in the future.
In conclusion, our five-center case-control study, for the first time, showed that METTL14 rs298982 G/A and rs1064034 T/A decreased the risk of ALL in southern Chinese children and suggested that the METTL14 polymorphism might be a potential biomarker for pediatric ALL susceptibility and chemotherapeutic choice. Nevertheless, these SNPs in the METTL14 gene are intriguing loci for further studies, and the underlying biological mechanisms should be revealed.
Abbreviations
METTL14, methyltransferase like 14; ALL, acute lymphoblastic leukemia; m6A, N6-methyladenosine; SNPs, single nucleotide polymorphisms; ORs, odds ratios; CIs confidence intervals; MRD, minimal residual disease; B-ALL, B-cell precursor acute lymphoblastic leukemia; T-ALL, T-cell lineages acute lymphoblastic leukemia; ARID5B, AT-rich interaction domain 5B; USP1, ubiquitin specific peptidase 1; METTL3, methyltransferase 3; WTAP, WT1 associated protein; FTO, fat mass and obesity-associated protein; ALKBH5, α-ketoglutarate-dependent dioxygenase homolog 5; SYSU, Sun Yat-sen University; GTEx, the Genotype-Tissue Expression; FPRP, false-positive report probability; AML, acute myelocytic leukemia; CCCG, Chinese Children Cancer Group chemotherapeutics; SCCLG, South China Children Leukemia Group chemotherapeutics; eQTL, expression quantitative trait loci; mRNA, messenger RNA; lncRNA, long noncoding RNA; SNHG8, small nucleolar RNA host gene 8.
Ethics Approval and Consent to Participate
The experiments were approved by the Ethics Committee of the Guangzhou Women and Children’s Medical Center. Written informed consent was obtained from all patients. The experiments were carried out following the Declaration of Helsinki.
Acknowledgments
We thank the Clinical Biological Resource Bank of Guangzhou Women and Children’s Medical Center for providing some of the clinical samples.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, agreed to the submitted journal, and agree to be accountable for all aspects of the work.
Funding
This work was supported by grants from the Natural Science Foundation of Guangdong Province (2020A1515010188), Medical Scientific Research Foundation of Guangdong Province, China (No. A 2019451) and National Natural Science Foundation of China (No. 81900150).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Mosaad YM, Khashaba M, Darwish A, et al. ARID5B rs10821936 and rs10994982 gene polymorphisms and acute lymphoblastic leukemia: relation to disease susceptibility and outcome. Pediatric hematology and oncology. 2019;36(6):365–375. doi:10.1080/08880018.2019.1649333
2. Abaji R, Gagne V, Xu CJ, et al. Whole-exome sequencing identified genetic risk factors for asparaginase-related complications in childhood ALL patients. Oncotarget. 2017;8(27):43752–43767. doi:10.18632/oncotarget.17959
3. Chen SS, Wu WZ, Zhang YP, Huang WJ. Gene polymorphisms of SOCS1 and SOCS2 and acute lymphoblastic leukemia. Eur Rev Med Pharmacol Sci. 2020;24(10):5564–5572. doi:10.26355/eurrev_202005_21342
4. Nordlund J, Syvanen AC. Epigenetics in pediatric acute lymphoblastic leukemia. Semin Cancer Biol. 2018;51:129–138. doi:10.1016/j.semcancer.2017.09.001
5. Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373(16):1541–1552. doi:10.1056/NEJMra1400972
6. Pui CH. Genomic and pharmacogenetic studies of childhood acute lymphoblastic leukemia. Front Med. 2015;9(1):1–9. doi:10.1007/s11684-015-0381-3
7. Yang X, Wu X, Fang N, et al. FOXO3 gene polymorphisms influence the risk of acute lymphoblastic leukemia in Chinese children. J Cell Biochem. 2020;121(2):2019–2026. doi:10.1002/jcb.29436
8. Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28(5):507–517. doi:10.1038/s41422-018-0034-6
9. He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18(1):176. doi:10.1186/s12943-019-1109-9
10. Dai D, Wang H, Zhu L, Jin H, Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018;9(2):124. doi:10.1038/s41419-017-0129-x
11. Weng H, Huang H, Wu H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22(2):191–205 e199. doi:10.1016/j.stem.2017.11.016
12. Sun C, Chang L, Liu C, Chen X, Zhu X. The study of METTL3 and METTL14 expressions in childhood ETV6/RUNX1-positive acute lymphoblastic leukemia. Mol Genet Genomic Med. 2019;7(10):e00933. doi:10.1002/mgg3.933
13. Zhuo Z, Lu H, Zhu J, et al. METTL14 gene polymorphisms confer neuroblastoma susceptibility: an eight-center case-control study. Mol Ther Nucleic Acids. 2020;22:17–26. doi:10.1016/j.omtn.2020.08.009
14. Liu X, Huang L, Huang K, et al. Novel associations between METTL3 gene polymorphisms and pediatric acute lymphoblastic leukemia: a five-center case-control study. Front Oncol. 2021;11:635251. doi:10.3389/fonc.2021.635251
15. He J, Yuan L, Lin H, et al. Genetic variants in m(6)A modification core genes are associated with glioma risk in Chinese children. Mol Ther Oncolytics. 2021;20:199–208. doi:10.1016/j.omto.2020.12.013
16. Hua RX, Zhuo ZJ, Zhu J, et al. Association between genetic variants in the XPG gene and gastric cancer risk in a Southern Chinese population. Aging. 2016;8(12):3311–3320. doi:10.18632/aging.101119
17. Zhuo ZJ, Liu W, Zhang J, et al. Functional polymorphisms at ERCC1/XPF genes confer neuroblastoma risk in Chinese children. EBioMedicine. 2018;30:113–119. doi:10.1016/j.ebiom.2018.03.003
18. Yang X, He J, Chang Y, et al. HOTAIR gene polymorphisms contribute to increased neuroblastoma susceptibility in Chinese children. Cancer. 2018;124(12):2599–2606. doi:10.1002/cncr.31353
19. Lin DY, Zeng D, Millikan R. Maximum likelihood estimation of haplotype effects and haplotype-environment interactions in association studies. Genet Epidemiol. 2005;29(4):299–312. doi:10.1002/gepi.20098
20. Hua RX, Zhuo Z, Ge L, et al. LIN28A gene polymorphisms modify neuroblastoma susceptibility: a four-centre case-control study. J Cell Mol Med. 2020;24(1):1059–1066. doi:10.1111/jcmm.14827
21. Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96(6):434–442. doi:10.1093/jnci/djh075
22. Ma JZ, Yang F, Zhou CC, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65(2):529–543. doi:10.1002/hep.28885
23. Chen X, Xu M, Xu X, et al. METTL14 suppresses CRC progression via regulating N6-methyladenosine-dependent primary miR-375 processing. Mol Ther. 2020;28(2):599–612. doi:10.1016/j.ymthe.2019.11.016
24. Sun T, Wu Z, Wang X, et al. LNC942 promoting METTL14-mediated m(6)A methylation in breast cancer cell proliferation and progression. Oncogene. 2020;39(31):5358–5372. doi:10.1038/s41388-020-1338-9
25. Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118(5):1590–1605. doi:10.1172/JCI34772
26. Fan D, Qiu B, Yang XJ, et al. LncRNA SNHG8 promotes cell migration and invasion in breast cancer cell through miR-634/ZBTB20 axis. Eur Rev Med Pharmacol Sci. 2020;24(22):11639–11649. doi:10.26355/eurrev_202011_23808
27. Tian X, Liu Y, Wang Z, Wu S. lncRNA SNHG8 promotes aggressive behaviors of nasopharyngeal carcinoma via regulating miR-656-3p/SATB1 axis. Biomed Pharmacother. 2020;131:110564. doi:10.1016/j.biopha.2020.110564
28. Qu X, Li Y, Wang L, Yuan N, Ma M, Chen Y. LncRNA SNHG8 accelerates proliferation and inhibits apoptosis in HPV-induced cervical cancer through recruiting EZH2 to epigenetically silence RECK expression. J Cell Biochem. 2020;121(10):4120–4129. doi:10.1002/jcb.29646
29. Miao W, Lu T, Liu X, Yin W, Zhang H. LncRNA SNHG8 induces ovarian carcinoma cells cellular process and stemness through Wnt/beta-catenin pathway. Cancer Biomarkers. 2020;28(4):459–471. doi:10.3233/CBM-190640
30. Dong J, Teng F, Guo W, Yang J, Ding G, Fu Z. lncRNA SNHG8 promotes the tumorigenesis and metastasis by sponging miR-149-5p and predicts tumor recurrence in hepatocellular carcinoma. Cell Physiol Biochem. 2018;51(5):2262–2274. doi:10.1159/000495871
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.