Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 13
Genetic variants in FAM13A and IREB2 are associated with the susceptibility to COPD in a Chinese rural population: a case-control study
Authors Zhang YN, Qiu J, Zhang P, Zhang J, Jiang M , Ma ZB
Received 11 January 2018
Accepted for publication 14 March 2018
Published 25 May 2018 Volume 2018:13 Pages 1735—1745
DOI https://doi.org/10.2147/COPD.S162241
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chunxue Bai
Yanan Zhang,1 Jie Qiu,1 Peng Zhang,1 Jin Zhang,1 Min Jiang,2 Zhanbing Ma3
1Department of Respiratory and Critical Care Medicine, General Hospital of Ningxia Medical University, Ningxia Medical University, Yinchuan, People’s Republic of China; 2National Engineering Research Center for Beijing Biochip Technology, Sub-center in Ningxia, General Hospital of Ningxia Medical University, Yinchuan, People’s Republic of China; 3Department of Medical Genetic and Cell Biology, Ningxia Medical University, Yinchuan, People’s Republic of China
Background: Genome-wide association studies identified several genomic regions associated with the risk of chronic obstructive pulmonary disease (COPD), including the 4q22 and 15q25 regions. These regions contain the FAM13A and IREB2 genes, which have been associated with COPD but data are lacking for Chinese patients. The objective of the study was to identify new genetic variants in the FAM13A and IREB2 associated with COPD in Northwestern China.
Methods: This was a case-control study performed in the Ningxia Hui Autonomous Region between January 2014 and December 2016. Patients were grouped as COPD and controls based on FEV1/FVC<70%. Seven tag single-nucleotide polymorphisms (SNPs) in the FAM13A and IREB2 genes were genotyped using the Agena MassARRAY platform. Logistic regression was used to determine the association between SNPs and COPD risk.
Results: rs17014601 in FAM13A was significantly associated with COPD in the additive (odds ratio [OR]=1.36, 95% confidence interval [CI]: 1.11–1.67, P=0.003), heterozygote (OR=1.76, 95% CI: 1.33–2.32, P=0.0001), and dominant (OR=1.67, 95% CI: 1.28–2.18, P=0.0001) models. Stratified analyses indicated that the risk was higher in never smokers. rs16969858 in IREB2 was significantly associated with COPD but in the univariate analysis only, and the multivariate analysis did not show any association.
Conclusion: The results suggest that the new variant rs17014601 in the FAM13A gene was significantly associated with COPD risk in a Chinese rural population. Additional studies are required to confirm the role of this variant in COPD development and progression.
Keywords: FAM13A, IREB2, chronic obstructive pulmonary disease, single-nucleotide polymorphism
Introduction
Chronic obstructive pulmonary disease (COPD) is a major cause of morbidity and mortality worldwide. It is characterized by persistent respiratory symptoms and airflow limitation.1 In People’s Republic of China, the prevalence of COPD in individuals ≥40 years of age is estimated at 8.2%2 or varying from 5% to 13% in different provinces/cities.3 Cigarette smoking is considered as the most important risk factor, but genetic characteristics play an important role in the susceptibility to COPD. Genome-wide association studies (GWAS) identified several genomic regions associated with higher COPD risk. Some GWAS loci are located in the FAM13A gene on chromosome 4q22 and in the 15q25 locus, which includes the IREB2 gene.4,5
GWAS showed that variants in FAM13A (family with sequence similarity 13, member A) were associated with FEV1/FVC and COPD.6–8 FAM13A was initially considered as a signal transduction gene because of the RhoGAP functional domain in the exon region,9 but it is now known to be associated with β-catenin signaling, which is typically activated during injury repair and tissue regeneration.10,11 Hypoxia commonly accompanies COPD and enhances FAM13A expression.9 In addition, Kim et al12 showed that FAM13A SNPs associated with a higher risk of COPD were also associated with an increased FAM13A expression in the lungs, suggesting a possible causative association with pathological changes in the lung. Corvol et al13 showed that the association of the FAM13A gene with pulmonary function parameters (FEV1%predicted and FEV1/FVC) was observed in different independent cohorts, suggesting that FAM13A is associated with a specific phenotype of COPD. Furthermore, Choo et al14 demonstrated an association between the CTGA diplotype in FAM13A and the emphysema phenotype of COPD, and Jiang et al11 provided the basis for the role of FAM13A in the development of emphysema. A recent study by Corvol et al15 showed that FAM13A and airway epithelial-mesenchymal transition (EMT) are closely associated in cystic fibrosis. EMT is also thought to play an important role in airway remodeling in COPD.16 Taken together, these results strongly suggest that FAM13A is involved in the etiology of lung diseases and COPD.
Iron is found in cigarette smoke17 and has been shown to disrupt the lung homeostasis, making lung tissues more susceptible to damage from any cause.18 IREB2 is a gene that is translated into the iron regulatory protein 2 (IRP2), which plays a key role in iron homeostasis. IREB2 is in strong linkage disequilibrium with nicotine receptor genes (CHRNA3 and 5).5 IREB2 expression is increased in the lungs of patients with COPD.19 IRP2 regulates cellular iron homeostasis and mitochondrial function.20,21 It is reported that some IREB2 variants may affect COPD in the presence of high levels of iron due to cigarette smoke exposure.18 Therefore, there could be some association between IREB2 and respiratory conditions such as COPD.
The association between single-nucleotide polymorphisms (SNPs) in FAM13A and IREB2 and the risk of COPD is still unclear, although some GWAS loci have been reported.4,5 We aimed to identify new genetic variants associated with COPD in People’s Republic of China. The aim of the present case-control study was to examine the association between tag SNPs22 in FAM13A and IREB2 and COPD risk. In addition, this is the first study evaluating the effect of genetic factors on the pathogenesis of COPD in the Ningxia Hui Autonomous Region (Northwest China).
Materials and methods
Study design and population
The COPD screening and early intervention project in the Ningxia Hui Autonomous Region was funded by the Ningxia government and aimed to carry out a prospective investigation between January 2014 and December 2016 to acquire data on COPD in the Ningxia Hui Autonomous Region. The investigation was approved by the ethics committee of the General Hospital of Ningxia Medical University. All participants provided a written informed consent.
A total of 6,130 participants ≥40 years of age and from a single township volunteered to participate in the study. They were local farmers, and at least three generations of their families were Han.
The participants were grouped as COPD and controls. COPD was defined as post-bronchodilator FEV1/FVC <70% and with chronic respiratory airway symptoms including dyspnea, chronic cough, sputum production, or wheezing. The exclusion criteria were as follows: 1) history of other respiratory diseases such as bronchial asthma, pulmonary tuberculosis, interstitial lung disease, or lung cancer or 2) unable to perform the lung function tests for any reason. The controls were with normal pulmonary function (FEV1/FVC >70%) and had no known medical illnesses or family disorders.
All volunteers underwent blood tests and completed questionnaires. The participants were interviewed by trained interviewers using standardized questionnaires about the risk factors of COPD. All pulmonary function measurements were performed using portable spirometers (MicroLab Spirometer, MD Spiro, Lewiston, ME, USA) and according to the guidelines of the American Thoracic Society.23 Peripheral blood samples (2 mL) were collected in EDTA Vacutainer tubes for DNA extraction.
SNP selection and genotyping
We selected seven tag SNPs in the FAM13A and IREB2 genes using the Genome Variation Server database (http://gvs.gs.washington.edu/GVS147/) and the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/projects/SNP) and based on the following criteria: tag SNPs in the CHB and Asian database selected by the Haploview 4.2 software (Broad Institute, Cambridge, MA, USA)24 with Hardy-Weinberg equilibrium (HWE) P-value ≥0.05, a minor allele frequency ≥0.05, and r2 ≥0.8.
Peripheral blood leukocyte DNA was extracted using a DNA extraction kit (Promega, Madison, WI, USA). DNA concentration was determined using a NanoDrop 1000 (Thermo Fisher Scientific, Waltham, MA, USA). Genotyping was done by CapitalBio Corporation (Beijing, People’s Republic of China) using a MassARRAY platform (Agena Biosciences Inc., San Diego, CA, USA), according to the manufacturer’s protocol. Multiplex reaction primers were designed using the MassARRAY Assay Design Tools on the Agena official website (https://agenacx.com/online-tools/) (Table 1). Mass determination was carried out using a MALDI-TOF mass spectrometer (Agena Biosciences Inc.), and the MassARRAY Type 4.0 software (Agena Biosciences Inc.) was used for data acquisition.
  | Table 1 SNPs and primers |
Covariate assessment
Demographic characteristics and COPD risk factors (age, sex, height, weight, smoking status, age at first cigarette, number of cigarettes/day, cooking and heating with coal stove, family history of lung disease, and childhood history of respiratory disease) were obtained using a questionnaire designed for this specific population. All participants were personally interviewed by trained interviewers.
The smoking status was defined as follows:25 subjects who had smoked >20 packs of cigarettes in a lifetime or 1 cigarette/d for a year were regarded as ever smokers (current or former); otherwise, they were classified as never smokers. Smokers who were still smoking at the time of the interview were considered as current smokers, and those who had quit (for at least 30 days before the interview) were former smokers.26 Pack-years were calculated in smokers by multiplying the average number of cigarettes smoked per day by the number of years of smoking and by dividing by 20 cigarettes/pack. Participants were classified as follows: 0, 0–20 and >20 pack-years. Cooking and heating with coal stove meant that the participants were using coal as domestic fuel. Body mass index (BMI) was calculated by dividing the weight (in kilograms) by the squared height (in meters).
Statistical analysis
Continuous data were presented as mean ± standard deviation, and categorical data were presented as frequency or percentage. The differences in the distributions of demographic characteristics, selected variables, and genotypes between the two groups were analyzed with Student’s t-test or the chi-square test, as appropriate. HWE for each SNP was tested using the chi-square test in the control group.
Unconditional logistic regression analyses without or with adjustment for the covariates (age, sex, BMI, smoking status, pack-years, coal consumption, pulmonary problems in childhood, and family history of pulmonary diseases) were used to estimate the odds ratio (OR) and 95% confidence interval (95% CI) for evaluating the effect of each SNP on COPD risk. Then, genetic variants were assessed using different genetic models (additive, dominant, and recessive). The subgroup analyses according to the abovementioned covariates were performed using stratified models.
All analyses were performed with R (http://www.R-project.org, The R Foundation) and Empowerstats (http://www.empowerstats.com, X&Y Solutions, Inc., Boston, MA, USA). Two-sided P-values <0.05 were considered statistically significant.
Results
Characteristics of the participants
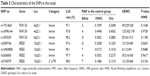
Figure 1 presents the study flowchart; 491 patients with COPD and 611 controls were included. Their characteristics are presented in Table 2. There were no differences in age, sex, coal use, and childhood pulmonary problems between the two groups (all P>0.05). Compared with the control group, the COPD group had significantly more smokers (P<0.0001), more pack-years smoked (P<0.0001), higher family history of pulmonary diseases (P=0.03), and lower BMI (P<0.0001). Among the patients with COPD, 25.9%, 45.8%, 21.6%, and 6.7% were classified as GOLD stages I, II, III, and IV, respectively, according to airflow limitation (FEV1%pre levels).
  | Figure 1 Study flowchart. |
  | Table 2 Characteristics of the subjects |
Association between SNPs and COPD susceptibility
The information about the selected SNPs is shown in Table 3. All of the SNPs were distributed within the parameters of HWE for the control population (P>0.05). The call rates during the genotyping of these SNPs were all above 95%.
The frequencies of the SNPs in the COPD and control groups are shown in Table 4. The frequencies of rs17014601 T>C and rs16969858 C>T were significantly different between the two groups (P=0.0005 and P=0.03, respectively). When considering the rs17014601 SNP in the FAM13A gene, compared with the TT genotype, the CT genotype was associated with an increased risk of COPD (non-adjusted OR=1.65, 95% CI=1.28–2.13). The association was stable in the age- and sex-adjusted analysis (adjusted OR=1.66, 95% CI=1.29–2.15), and even after adjusting for more covariates (age, sex, BMI, smoking status, pack-years smoked, coal use, family history of lung disease, and childhood history of respiratory disease) (adjusted OR=1.76, 95% CI=1.33–2.32).
Five genetic models (additive, heterozygote, homozygote, dominant, and recessive) were used to analyze the associations between the variants and risk of COPD with and without adjustments for covariates (Table 5). The rs17014601 T>C SNP fitted the additive model, showing a significantly increased risk of COPD in the presence of a C allele, in a dose-dependent manner, and after adjustment for age, sex, and other covariates (adjusted OR=1.36, 95% CI=1.11–1.67, P=0.0028). The rs17014601 CT genotype was associated with an increased risk of COPD based on the heterozygote model (adjusted OR=1.76; 95% CI=1.33–2.32; P=0.0001) and dominant model (adjusted OR=1.67; 95% CI=1.28–2.18; P=0.0001).
No associations between the other SNPs and COPD were observed in the multivariate logistic regression analyses and genetic models.
Stratification analysis
The association between variant genotypes and COPD risk was further evaluated using analyses stratified by age, sex, BMI, smoking status, pack-years smoked, coal use, family history of lung disease, and childhood history of respiratory disease (Table 6). The FAM13A rs17014601 CT genotype was associated with an increased COPD risk in never smokers (adjusted OR=1.97, 95% CI=1.05–3.72, P=0.0356) and 0 pack-years smoked (adjusted OR=2.06, 95% CI=1.44–2.96, P=0.0001), when compared to the respective reference groups.
Stratified analysis of association between the rs16969858 SNP and COPD risk provided no statistically significant result (Table S1). There was no effect of the rs16969858 SNP genotype in different smoking status (Table S2). There was no effect of the rs16969858 SNP genotype in different smoking status according to the dominant and recessive models (Table S3). There was no effect of the rs17014601 SNP in different smoking statuses (Table S4), nor according to the dominant and recessive models (Table S5).
Discussion
In the present population-based case-control study, we investigated the potential associations of FAM13A rs17014601 T>C, rs16996144 G>A, and rs1870339 C>G and IREB2 rs2009746 A>G, rs16969858 C>T, rs2656065 G>A, and rs3743079 C>T SNPs with COPD susceptibility in a Chinese rural population. We found that FAM13A rs17014601 had an independent effect on the COPD risk. The C allele in the FAM13A rs17014601 was significantly associated with COPD risk or occurrence in a Chinese rural population.
FAM13A on chromosome 4q22 has been consistently associated with COPD by GWAS. Among many tested SNPs, the rs7671167 SNP in FAM13A is most highly associated with COPD in Caucasians7,27,28 and Asians, especially in Chinese.29 Indeed, Xie et al29 confirmed that the FAM13A rs7671167 SNP was associated with COPD risk in a Chinese Han population and that rs7671167 was related to lung function decline. In addition, Guo et al30 reported that the frequency of the rs2869967 C allele was significantly increased in Chinese patients with COPD,30 and Wang et al31 showed that five FAM13A SNPs (rs7671167, rs2869966, rs2869967, rs2045517, and rs6830970) were associated with the FEV1/FVC ratio in all subjects and that rs6830970 was associated with the FEV1/FVC ratio in the COPD subset. Furthermore, the FAM13A locus is apparently not influenced by smoking.4 van der Plaat et al32 suggested that the FAM13A loci (including rs6849143) are significantly associated with lung function measurements in never smokers.
In the present study, we found that a new FAM13A variant contributed to COPD risk in a Chinese rural population. Indeed, rs17014601 was significantly associated with an increased risk of COPD. In People’s Republic of China, COPD is more common among rural residents compared with urban residents,2 probably because of a number of environmental risk factors such as old age, smoking, coal use, infection, and low body mass index. Ningxia is an agricultural region in Northwestern China. Coal stove cooking and winter coal heating are very common. One advantage of our study is that we conducted an investigation of the possible COPD risk factors in individuals who were not yet treated for any lung disease. Then, we adjusted these associations according to known risk factors for COPD, including coal use. The association between rs17014601 and COPD remained significant after adjusting for the confounding factors. In addition, stratified analyses showed that never smokers had a higher risk of COPD in association with FAM13A rs17014601 compared with the whole cohort. This is the first report of the association between the FAM13A rs17014601 and COPD risk, but the results have to be confirmed by additional studies.
The exact biological functions of FAM13A are still unknown. The RhoGAP domain in the exon region may be related to COPD.13 Rho GTPases are key regulators of cytoskeletal dynamics involved in pulmonary endothelial barrier functions, and have been shown to be dysregulated in several lung diseases.33 It is probable that genetic variations of FAM13A may affect Rho GTPases activity and the cellular pathways associated with FAM13A, hereby contributing to lung disease. The most significant SNPs in FAM13A have been found in non-coding regions downstream of the RhoGAP domain and are associated with FAM13A gene expression levels.7 Recently, the biological function of FAM13A in emphysema development has been explored: Jiang et al11 reported the expression of the FAM13A protein in airways, alveolar epithelial cells, and alveolar macrophages. FAM13A knockout mouse models are less susceptible to develop emphysema.11 In vitro experiments showed that FAM13A interacts with PP2A and promotes the degradation of β-catenin in bronchial epithelial cells, inhibiting the activation of the Wnt pathway.10 Moreover, metabolic regulation may be another mechanism by which FAM13A promotes CS-induced emphysema.34 Indeed, FAM13A promotes fatty acid oxidation (FAO) and subsequent increases in ROS, possibly by interacting with Sirtuin 1 (SIRT1) and increasing the expression of CPT1A, a key mitochondrial enzyme for the FAO pathway, thereby enhancing FAO. These findings suggest that the impact of FAM13A on COPD could be independent from smoke exposure.
A previous study in Caucasians showed that IREB2 polymorphisms had an effect of COPD susceptibility, independently from smoking.27 A Russian study showed associations between the rs13180 SNP in IREB2 and COPD and lung function in the Tatar population.35 A study from Poland suggests that the rs2568494 SNP in IREB2 was not associated with COPD, but with lung cancer.28 In the present study, the rs16969858 SNP in the IREB2 gene was significantly associated with COPD only in the univariate analysis, and multivariate analysis did not show any association. These discrepancies may be due to the selection of the SNPs being studied, as well as to the interactions with other environmental and genetic factors. Indeed, the rs16969858 SNP is an intronic SNP that could cause dysfunction because of differential splicing. A recent genome-wide bioinformatics study suggested that there were no differences in gene expression of IREB2 and FAM13A, but that the expression of these genes was dependent on the interactions of genes.36 This suggests new research avenues and expression studies in lung tissues should be performed.
There are some limitations in the present study that should be addressed. The number of COPD cases was limited. Larger scale studies are needed in different populations to validate the result. Although we were able to identify associations, we were not able to identify the causal mechanisms. This is the first study on the association between rs17014601 and COPD. The Han ethnicity was selected because it is the major ethnic group in China. Nevertheless, we agree that other ethnic groups will have to be studied. In addition, the Han populations that were enrolled in the COPD Susceptibility Study were mainly from southern China. In the present study, we included Chinese Han people from Northwest China and it is possible that there are genetic differences between South and North China.37 Additional studies are still necessary to understand the genetics of COPD.
Conclusion
The present study strongly suggests an association between the FAM13A gene and COPD in Chinese rural patients. Further studies are required to elucidate the functional roles of these variants, which may have an impact on the management of lung diseases.
Acknowledgments
We would like to thank the individuals who participated in this study, as well as the staff who helped complete the questionnaires and collect blood samples: Xiulan Wu, Zhanfu Yao, and Zhonggang Zhao (from the Center for Disease Control and Prevention in Qingtongxia city, Ningxia Hui Autonomous Region, People’s Republic of China); Xingyan Lu and Liankai Chen (from the Ye Sheng town hospital in Qingtongxia City, Ningxia Hui Autonomous Region, People’s Republic of China); and Tao Ma (from the Qingtongxia City Hospital, Ningxia Hui Autonomous Region, People’s Republic of China). This study was funded by the Ningxia Science and Technology Huimin project (Grant No 2014KJHM05), the Ningxia Medical University scientific research project (Grant No 2001210203), and the National Natural Science Foundation of China (Grant No 81760011).
Disclosure
The authors report no conflicts of interest in this work.
References
GOLD. Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Pulmonary Disease – 2018 Report. Global Initiative for Chronic Obstructive Lung Disease, Inc.; 2018. Available from: http://goldcopd.org/. Accessed May 02, 2018. | ||
Zhong N, Wang C, Yao W, et al. Prevalence of chronic obstructive pulmonary disease in China: a large, population-based survey. Am J Respir Crit Care Med. 2007;176:753–760. | ||
Fang X, Wang X, Bai C. COPD in China: the burden and importance of proper management. Chest. 2011;139(4):920–929. | ||
Pillai SG, Kong X, Edwards LD, et al; ECLIPSE and ICGN Investigators. Loci identified by genome-wide association studies influence different disease-related phenotypes in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182(12):1498–1505. | ||
Hardin M, Zielinski J, Wan ES, et al. CHRNA3/5, IREB2, and ADCY2 are associated with severe chronic obstructive pulmonary disease in Poland. Am J Respir Cell Mol Biol. 2012;47(2):203–208. | ||
Hancock DB, Eijgelsheim M, Wilk JB, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42(1):45–52. | ||
Cho MH, Boutaoui N, Klanderman BJ, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42(3):200–202. | ||
Cho MH, Castaldi PJ, Hersh CP, et al; NETT Genetics, ECLIPSE, and COPDGene Investigators. A genome-wide association study of emphysema and airway quantitative imaging phenotypes. Am J Respir Crit Care Med. 2015;192(5):559–569. | ||
Cohen M, Reichenstein M, Everts-van der Wind A, et al. Cloning and characterization of FAM13A1 – a gene near a milk protein QTL on BTA6: evidence for population-wide linkage disequilibrium in Israeli Holsteins. Genomics. 2004;84(2):374–383. | ||
Jin Z, Chung JW, Mei W, et al. Regulation of nuclear-cytoplasmic shuttling and function of Family with sequence similarity 13, member A (Fam13a), by B56-containing PP2As and Akt. Mol Biol Cell. 2015;26(6):1160–1173. | ||
Jiang Z, Lao T, Qiu W, et al. A chronic obstructive pulmonary disease susceptibility gene, FAM13A, regulates protein stability of β-catenin. Am J Respir Crit Care Med. 2016;194(2):185–197. | ||
Kim WJ, Lim MN, Hong Y, et al. Association of lung function genes with chronic obstructive pulmonary disease. Lung. 2014;192(4):473–480. | ||
Corvol H, Hodges CA, Drumm ML, Guillot L. Moving beyond genetics: is FAM13A a major biological contributor in lung physiology and chronic lung diseases? J Med Genet. 2014;51(10):646–649. | ||
Choo JY, Lee KY, Shin C, et al. Quantitative analysis of lungs and airways with CT in subjects with the chronic obstructive pulmonary disease (COPD) candidate FAM13A gene: case control study for CT quantification in COPD risk gene. J Comput Assist Tomogr. 2014;38(4):597–603. | ||
Corvol H, Rousselet N, Thompson KE, et al. FAM13A is a modifier gene of cystic fibrosis lung phenotype regulating rhoa activity, actin cytoskeleton dynamics and epithelial-mesenchymal transition. J Cyst Fibros. 2018;17(2):190–203. | ||
Bartis D, Mise N, Mahida RY, Eickelberg O, Thickett DR. Epithelial-mesenchymal transition in lung development and disease: does it exist and is it important? Thorax. 2014;69(8):760–765. | ||
Khiroya H, Turner AM. The role of iron in pulmonary pathology. Multidiscip Respir Med. 2015;10:34. | ||
Ghio AJ, Hilborn ED, Stonehuerner JG, et al. Particulate matter in cigarette smoke alters iron homeostasis to produce a biological effect. Am J Respir Crit Care Med. 2008;178(11):1130–1138. | ||
Kim WJ, Lee SD. Candidate genes for COPD: current evidence and research. Int J Chron Obstruct Pulmon Dis. 2015;10:2249–2255. | ||
Sanchez M, Galy B, Schwanhaeusser B, et al. Iron regulatory protein-1 and −2: transcriptome-wide definition of binding mRNAs and shaping of the cellular proteome by iron regulatory proteins. Blood. 2011;118(22):e168–e179. | ||
Cloonan SM, Glass K, Laucho-Contreras ME, et al. Mitochondrial iron chelation ameliorates cigarette smoke-induced bronchitis and emphysema in mice. Nat Med. 2016;22(2):163–174. | ||
Nicolas P, Sun F, Li LM. A model-based approach to selection of tag SNPs. BMC Bioinformatics. 2006;7:303. | ||
Miller MR, Hankinson J, Brusasco V, et al; ATS/ERS Task Force. Standardisation of spirometry. Eur Respir J. 2005;26(2):319–338. | ||
Barrett JC. Haploview: visualization and analysis of SNP genotype data. Cold Spring Harb Protoc. 2009;2009(10):pdb ip71. | ||
Lamprecht B, McBurnie MA, Vollmer WM, et al; BOLD Collaborative Research Group. COPD in never smokers: results from the population-based burden of obstructive lung disease study. Chest. 2011;139(4):752–763. | ||
Kianoush S, Bittencourt MS, Lotufo PA, et al. Association between smoking and serum GlycA and high-sensitivity C-reactive protein levels: the multi-ethnic study of atherosclerosis (MESA) and Brazilian longitudinal study of adult health (ELSA-Brasil). J Am Heart Assoc. 2017;6(8):e006545. | ||
Siedlinski M, Tingley D, Lipman PJ, et al; COPDGene and ECLIPSE Investigators. Dissecting direct and indirect genetic effects on chronic obstructive pulmonary disease (COPD) susceptibility. Hum Genet. 2013;132(4):431–441. | ||
Ziólkowska-Suchanek I, Mosor M, Gabryel P, et al. Susceptibility loci in lung cancer and COPD: association of IREB2 and FAM13A with pulmonary diseases. Sci Rep. 2015;5:13502. | ||
Xie J, Wu H, Xu Y, et al. Gene susceptibility identification in a longitudinal study confirms new loci in the development of chronic obstructive pulmonary disease and influences lung function decline. Respir Res. 2015;16:49. | ||
Guo Y, Lin H, Gao K, et al. Genetic analysis of IREB2, FAM13A and XRCC5 variants in Chinese Han patients with chronic obstructive pulmonary disease. Biochem Biophys Res Commun. 2011;415(2):284–287. | ||
Wang B, Liang B, Yang J, et al. Association of FAM13A polymorphisms with COPD and COPD-related phenotypes in Han Chinese. Clin Biochem. 2013;46(16–17):1683–1688. | ||
van der Plaat DA, de Jong K, Lahousse L, et al. Genome-wide association study on the FEV1/FVC ratio in never-smokers identifies HHIP and FAM13A. J Allergy Clin Immunol. 2017;139(2):533–540. | ||
Duluc L, Wojciak-Stothard B. Rho GTPases in the regulation of pulmonary vascular barrier function. Cell Tissue Res. 2014;355(3):675–685. | ||
Jiang Z, Knudsen NH, Wang G, et al. Genetic control of fatty acid β-oxidation in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2017;56(6):738–748. | ||
Korytina GF, Akhmadishina LZ, Viktorova EV, Kochetova OV, Viktorova TV. IREB2, CHRNA5, CHRNA3, FAM13A and hedgehog interacting protein genes polymorphisms and risk of chronic obstructive pulmonary disease in Tatar population from Russia. Indian J Med Res. 2016;144(6):865–876. | ||
Morrow JD, Zhou X, Lao T, et al. Functional interactors of three genome-wide association study genes are differentially expressed in severe chronic obstructive pulmonary disease lung tissue. Sci Rep. 2017;7:44232. | ||
Xu S, Yin X, Li S, et al. Genomic dissection of population substructure of Han Chinese and its implication in association studies. Am J Hum Genet. 2009;85(6):762–774. |
Supplementary materials
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.