Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 14
Genetic Polymorphisms of FCRL3, NLRP3 and IL2 are Associated with the Risk of Head and Neck Cancer in a Chinese Population
Received 14 June 2021
Accepted for publication 6 August 2021
Published 26 August 2021 Volume 2021:14 Pages 1047—1053
DOI https://doi.org/10.2147/PGPM.S324750
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Yuhao Zhang,1,2 Dawei Sun3
1Department of Otolaryngology Head and Neck Surgery, Shanxi Bethune Hospital, Shanxi Academy of Medical Sciences, Tongji Shanxi Hospital, Third Hospital of Shanxi Medical University, Taiyuan, 030032, People’s Republic of China; 2Department of Otolaryngology Head and Neck Surgery, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430030, People’s Republic of China; 3Department of Thyroid Surgery, The Affiliated Hospital of Qingdao University, Qingdao, Shandong, 266000, People’s Republic of China
Correspondence: Dawei Sun
Department of Thyroid Surgery, The Affiliated Hospital of Qingdao University, #16 Jiangsu Road, Qingdao, Shandong, 266000, People’s Republic of China
Email [email protected]
Purpose: This study aimed to evaluate the associations between immune-related gene (FCRL3, NLRP3 and IL2) polymorphisms and the risk of head and neck cancer (HNC).
Methods: Six polymorphisms of FCRL3, NLRP3 and IL2 were genotyped in 400 HNC cases and 400 controls using a MassARRAY platform.
Results: rs11264799-T was a protective variant against HNC risk, while rs7528684-G, rs35829419-A and rs6822844-T were all risk alleles for HNC (p < 0.05). rs11264799-TT was correlated with reduced HNC risk, while rs7528684-GG and rs6822844-TG were associated with an elevated risk of disease (p < 0.05). Moreover, rs11264799 was correlated with a declining risk of HNC in three genetic models (p < 0.05). In contrast, rs7528684 exhibited an elevated risk of HNC in recessive and additive models; rs35829419 and rs6822844 were associated with an increased risk of disease in dominant and additive models (p < 0.05). Finally, an interaction was observed between the above SNPs and drinking (p < 0.05).
Conclusion: The results expand our knowledge on immune-related gene polymorphisms in HNC and provide clues for further functional study on the pathogenesis of HNC.
Keywords: head and neck cancer, HNC, Fc receptor-like 3, FCRL3, NLR family pyrin domain containing 3, NLRP3, interleukin 2, IL2, genetic polymorphisms
Introduction
Head and neck cancer (HNC) ranks as the sixth most common type of cancer worldwide, with an estimated 500,000 newly diagnosed cases and 325,000 deaths annually.1 The primary sites of HNC mainly include the pharynx, larynx and oral cavity. In recent years, although specific medicine and novel treatment approaches have improved the prognosis of HNC patients to a great extent, it is still necessary to find other ways to decrease the incidence of disease.2 Among researchers, it is believed that HNC is a complicated disease affected by both gene polymorphisms and environmental factors such as smoking and drinking.3 Previous studies have identified a number of variants associated with HNC risk, mainly including single-nucleotide polymorphisms (SNPs) in drug and alcohol metabolism- and cell cycle control-related genes.4 However, the current data represent only a small part of the genetic predisposition to HNC, and there is still an urgent need to identify more novel susceptible SNPs.
Recent clinical trials have revealed that immunotherapy is a promising treatment approach for advanced HNC patients,5 indicating that immune-related genes may play important roles in the progression of HNC. Fc receptor-like 3 (FCRL3) is a member of the FCRL superfamily and is involved in the regulation of the immune system.6 Polymorphisms of FCRL3 have been identified as taking part in the initiation of a diversity of autoimmune diseases,7 whereas little information has been found regarding the contribution of its polymorphisms to HNC risk. NLR family pyrin domain containing 3 (NLRP3) has specific structural domains that form the inflammasome complex NALP3 and are thus involved in the mediation of inflammation and the immune response in the human body.8 Moreover, interleukin 2 (IL2) is an important cytokine that is crucial for the proliferation of T and B cells, and its polymorphisms are associated with various cancers.9 Thus, FCRL3, NLRP3 and IL2 were selected as candidate genes in our study.
Based on previous association studies, six SNPs in FCRL3, NLRP3 and IL2 were selected for investigation. rs11264799 and rs7528684 in FCRL3 have been associated with the progression of multiple autoimmune diseases.10 rs35829419 in NLRP3 has been identified as a risk factor for the occurrence and outcome of colorectal cancer.11 rs10754558 and rs4353135 have been investigated in previous studies on gastric cancer and immune disorders, respectively.12,13 In addition, rs6822844 in IL2 has been proven to be associated with plasma IL2 levels in breast cancer patients, making it a promising biomarker for immune status among cancer patients.14
Considering the above associations between these SNPs and cancer and immune disorders, we speculated that these SNPs may also be involved in the occurrence and development of HNC. In this study, we genotyped the above six SNPs in a case-control cohort and evaluated their association with HNC risk, aiming to better understand the genetic predisposition to HNC.
Materials and Methods
Subjects
The present case-control study consists of 400 histopathologically diagnosed HNC cases and 400 healthy controls. The participants were all Chinese Han individuals recruited at the Affiliated Hospital of Qingdao University. All cases were newly diagnosed and previously untreated. The controls were blood donors without a history of cancer, immune disorder or serious disease. We obtained written informed consent from each subject, and the study was approved by the Ethics Department of the Affiliated Hospital of Qingdao University (No. 201204–13) and was carried out in accordance with the World Medical Association Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects.
Genotyping
Five milliliters of whole blood was collected from each subject in tubes containing ethylenediaminetetraacetic acid. DNA was extracted using a PureLink™ Pro 96 Genomic DNA Purification Kit (Invitrogen, Carlsbad, CA). Primers were designed using Sequenom MassARRAY Assay Design 3.0 software. Genotypes were detected by Sequenom MassARRAY RS1000 (Sequenom, San Diego, CA).
Statistical Analyses
Statistical analyses were performed with SPSS package version 20.0 (SPSS, Chicago, IL, USA). Minor allele frequencies (MAFs) in controls were checked for departure from Hardy–Weinberg equilibrium (HWE) in controls. The associations between SNPs and HNC risk were evaluated using SNPstats (https://www.snpstats.net/start.htm) and are expressed as odds ratios (ORs) and 95% confidence intervals (CIs). Statistical significance was established when p < 0.05.
Results
The basic characteristics of the participants are presented in Table 1, including sex, age, and smoking and drinking status. No significant differences were observed in the distributions of sex and age between the case and control groups, suggesting that the two groups were comparable (p > 0.05). However, there were significantly more smokers and drinkers than in the controls, suggesting that tobacco and alcohol consumption might be risk factors (p < 0.05). The primary sites of HNC cases were 46.75% in the pharynx, 30.50% in the larynx and 22.75% in the oral cavity.
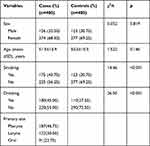 |
Table 1 Basic Characteristics of the Participants |
The basic information and MAFs of candidate SNPs are listed in Table 2. All of the SNPs were consistent with HWE (p > 0.05). We found that four SNPs had a significant allelic frequency distribution between the case and control groups. The MAF of rs11264799 was lower in cases than in controls, making it a protective variant against HNC risk (OR = 0.761, 95% CI: 0.615–0.943, p = 0.012). In contrast, the MAF of rs7528684 in HNC patients was more prominent than in healthy donors, hinting that rs7528684-G might be a risk allele for HNC (OR = 1.260, 95% CI: 1.035–1.533, p = 0.021). Additionally, rs35829419-A (OR = 1.632, 95% CI: 1.152–2.311, p = 0.005) and rs6822844-T (OR = 1.600, 95% CI: 1.126–2.275, p = 0.008) were both risk alleles for HNC.
 |
Table 2 Allele Frequency Distributions Among HNC Cases and Healthy Controls |
The genotypic frequencies of candidate SNPs were also compared between cases and controls (Table 3). The frequency of rs11264799-TT in HNC cases was lower; thus, rs11264799-TT was correlated with a reduced risk of HNC (OR = 0.54, 95% CI: 0.33–0.89, p = 0.031), while rs7528684-GG (OR = 1.63, 95% CI: 1.10–2.43, p = 0.044) and rs6822844-TG (OR = 1.75, 95% CI: 1.18–2.60, p = 0.010) were found to be more prominent in HNC cases than in controls, which made them associated with an elevated risk of HNC.
 |
Table 3 Genotype Frequency Distributions Among HNC Cases and Healthy Controls |
Three classic genetic models were used to evaluate associations between SNPs and disease (Table 4). One protective variant and three risk variants were identified in different models. rs11264799 was correlated with a declining risk of HNC in all three models (p < 0.05). In contrast, rs7528684 exhibited an elevated risk of HNC in recessive and additive models, and rs35829419 and rs6822844 were associated with an increased risk of disease in dominant and additive models (p < 0.05).
 |
Table 4 Associations Between Candidate SNPs and HNC Risk in Three Genetic Models |
Finally, the interactions of four polymorphisms and tobacco and alcohol consumption were evaluated using a stratification analysis (Table 5). The effects of four SNPs on the risk of HNC were not affected by smoking status (p < 0.05). However, when participants were stratified by drinking status, the four SNPs had associations with HNC risk only in drinkers (p < 0.05).
 |
Table 5 Associations of Candidate SNPs with HNC Risk in Four Subgroups |
Discussion
Identification of genetic and environmental risk factors is not only helpful for us to better understand the pathogenesis of HNC but also beneficial to decrease the incidence of disease from the original. Immune-related genes have been extensively investigated in association studies on cancer. In this study, we genotyped six SNPs in the immune-related genes FCRL3, NLRP3 and IL2 and found that rs11264799 was a protective factor against HNC risk, while rs7528684, rs35829419, and rs6822844 were associated with an increased HNC risk.
FCRL3 is an immunoglobulin receptor and has specific immunoreceptor-tyrosine activation/inhibitor motifs in its cytoplasmic domain, affecting the modulation of the immune response.15 Genetic variation in FCRL3 has been associated with a wide range of diseases in different samples, including allergic rhinitis,16 sudden sensorineural hearing loss,17 autoimmune thyroid disease,18 rheumatoid arthritis,19 and multiple sclerosis.20 rs11264799 and rs7528684 in FCRL3 have been investigated in many association studies, and a meta-analysis has verified a significant association between these two SNPs and human autoimmune diseases.21 In the present study, we demonstrated novel correlations between rs11264799 and rs7528684 and HNC risk: rs11264799-T may protect carriers from HNC risk, while rs7528684-G contributes to an elevated risk of disease. rs11264799 and rs7528684 are located in the 5ʹ UTR and upstream transcript region of FCRL3, respectively, meaning that their polymorphisms may change the expression levels of FCRL3 and other related downstream genes. Thus, we speculated that rs11264799 and rs7528684 may exert an influence on the predisposition to HNC by changing the molecular function of FCRL3 in HNC patients; however, this hypothesis still needs to be further confirmed.
NLRP3 is a pyrin-like protein that can interact with the cell apoptosis-related protein PYCARD/ASC and from the NALP3 inflammasome complex (an upstream inducer of NF-kappaB signaling) with other proteins.22 Therefore, NLRP3 plays important roles in cell apoptosis and the immune response. Polymorphisms of NLRP3 have been associated with a diversity of diseases, such as Parkinson’s disease,23 Alzheimer’s disease,24 insulin resistance,25 ischemic stroke26 and other autoimmune and inflammatory diseases.27 rs35829419 is a missense variant (Q705K) in NLRP3 and has been identified as a susceptible risk factor for multiple human diseases.28 However, little evidence is available to determine its role in HNC. Our study demonstrated a significant correlation between rs35829419 and HNC risk by a comprehensive evaluation (allele, genotype frequency, and genetic model analysis). We suggested that carriers with the rs35829419-A allele should focus on their health: keeping away from tobacco and alcohol, maintaining a balanced diet and exercise, and having periodic physical examinations will reduce the risk of HNC.
IL2 is a secreted cytokine that is mainly produced by T cells. IL2 can promote the proliferation and activation of T, B and NK cells, consequently participating in the immune response. The effects of IL2 polymorphisms on the risk of cancer have been extensively investigated, including esophageal, gastric, hepatocellular, breast and bladder cancers.29 A recent study revealed that rs6822844 in IL2 was associated with plasma IL2 levels and might be used as a blood-based biomarker in the evaluation of immune status among breast cancer patients.14 Our study found that rs6822844-T contributed to a 1.6-fold increased risk of HNC, suggesting that rs6822844-T may also alter the expression of IL2 and impair the normal function of the immune system of HNC patients. However, the detailed mechanism needs to be explored in further studies.
Given that smoking and drinking are certain risk factors for HNC, we verified the associations between rs11264799, rs7528684, rs35829419, rs6822844 and HNC risk by stratification analysis. The associations were significant in both smokers and nonsmokers, suggesting that these SNPs are independent protective/risk factors for HNC that are not affected by smoking. However, although four SNPs had effects on the risk of disease in drinkers, no significant correlation was observed in nondrinkers. We speculated that there might be an interaction between these SNPs and drinking in the development of HNC.
In conclusion, we found that rs11264799 contributed to a lower HNC risk, while rs7528684, rs35829419, rs6822844 were associated with an increased HNC risk. The results expand our knowledge on immune-related gene polymorphisms in HNC and provide clues for further functional study on the pathogenesis of HNC.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, agreed to the submitted journal, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Lee YCA, Li S, Chen Y, et al. Tobacco smoking, alcohol drinking, betel quid chewing, and the risk of head and neck cancer in an East Asian population. Head Neck. 2019;41(1):92–102.
2. Guo W, Song H. Development of gene therapeutics for head and neck cancer in china: from bench to bedside. Hum Gene Ther. 2018;29(2):180–187. doi:10.1089/hum.2017.230
3. Kawakita D, Matsuo K. Alcohol and head and neck cancer. Cancer and Metastasis Reviews. 2017;36(3):425–434. doi:10.1007/s10555-017-9690-0
4. Cadoni G, Boccia S, Petrelli L, et al. A review of genetic epidemiology of head and neck cancer related to polymorphisms in metabolic genes, cell cycle control and alcohol metabolism. ACTA otorhinolaryngologica italica. 2012;32(1):1–11.
5. Mandal R, Şenbabaoğlu Y, Desrichard A, et al. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight. 2016;1(17):e89829. doi:10.1172/jci.insight.89829
6. Agarwal S, Kraus Z, Dement-Brown J, Alabi O, Starost K, Tolnay M. Human Fc Receptor-like 3 Inhibits Regulatory T Cell Function and Binds Secretory IgA. Cell Rep. 2020;30(5):1292–1299. doi:10.1016/j.celrep.2019.12.099
7. Kochi Y, Yamada R, Suzuki A, et al. A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several autoimmunities. Nat Genet. 2005;37(5):478–485. doi:10.1038/ng1540
8. He Y, Hara H, Núñez G. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem Sci. 2016;41(12):1012–1021. doi:10.1016/j.tibs.2016.09.002
9. Rogers O, Yen H, Solomon A, Drake C, Denmeade S. An IL‐2 proaerolysin fusion toxin that selectively eliminates regulatory t cells to enhance antitumor immune response. Prostate. 2019;79(10):1071–1078. doi:10.1002/pros.23819
10. Li J, Ma S, Shao L, et al. Inflammation-related gene polymorphisms associated with primary immune thrombocytopenia. Front Immunol. 2017;8:744. doi:10.3389/fimmu.2017.00744
11. Ungerbäck J, Belenki D, Jawad ul-Hassan A, et al. Genetic variation and alterations of genes involved in NFκB/TNFAIP3-and NLRP3-inflammasome signaling affect susceptibility and outcome of colorectal cancer. Carcinogenesis. 2012;33(11):2126–2134. doi:10.1093/carcin/bgs256
12. Castano-Rodriguez N, Kaakoush NO, Goh K-L, Fock KM, Mitchell HM. The NOD-like receptor signalling pathway in Helicobacter pylori infection and related gastric cancer: a case-control study and gene expression analyses. PLoS One. 2014;9(6):e98899. doi:10.1371/journal.pone.0098899
13. Yang C-A, Chiang B-L. Inflammasomes and human autoimmunity: a comprehensive review. J Autoimmun. 2015;61:1–8. doi:10.1016/j.jaut.2015.05.001
14. Lewin NL, Luetragoon T, Andersson B-Å, et al. The influence of single nucleotide polymorphisms and adjuvant radiotherapy on systemic inflammatory proteins, chemokines and cytokines of patients with breast cancer. Anticancer Res. 2019;39(3):1287–1292. doi:10.21873/anticanres.13240
15. Chistiakov DA, Chistiakov AP. Is FCRL3 a new general autoimmunity gene? Hum Immunol. 2007;68(5):375–383. doi:10.1016/j.humimm.2007.01.013
16. Gu Z, Hong S-L, Ke X, et al. FCRL3 gene polymorphisms confer autoimmunity risk for allergic rhinitis in a Chinese Han population. PLoS One. 2015;10(1):e0116419. doi:10.1371/journal.pone.0116419
17. Liu H, Gu Z, Kang H-Y, et al. FCRL3 gene polymorphisms confer risk for sudden sensorineural hearing loss in a Chinese Han Population. Gene. 2015;570(1):89–94. doi:10.1016/j.gene.2015.06.005
18. Inoue N, Watanabe M, Yamada H, et al. Associations between autoimmune thyroid disease prognosis and functional polymorphisms of susceptibility genes, CTLA4, PTPN22, CD40, FCRL3, and ZFAT, previously revealed in genome-wide association studies. J Clin Immunol. 2012;32(6):1243–1252. doi:10.1007/s10875-012-9721-0
19. Lin X, Zhang Y, Chen Q. FCRL3 gene polymorphisms as risk factors for rheumatoid arthritis. Hum Immunol. 2016;77(2):223–229. doi:10.1016/j.humimm.2015.12.007
20. Matesanz F, Fernández O, Milne RL, et al. The high producer variant of the Fc-receptor like-3 (FCRL3) gene is involved in protection against multiple sclerosis. J Neuroimmunol. 2008;195(1–2):146–150. doi:10.1016/j.jneuroim.2008.01.004
21. Yang Y, Su X, Zhang K, Zhou R. The Fc receptor-like 3 gene polymorphisms and susceptibility to autoimmune diseases: an updated meta-analysis. Autoimmunity. 2013;46(8):547–558. doi:10.3109/08916934.2013.835804
22. Shimada K, Crother TR, Karlin J, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36(3):401–414. doi:10.1016/j.immuni.2012.01.009
23. von Herrmann KM, Salas LA, Martinez EM, et al. NLRP3 expression in mesencephalic neurons and characterization of a rare NLRP3 polymorphism associated with decreased risk of Parkinson’s disease. NPJ Parkinson’s Dis. 2018;4(1):1–9.
24. Tan M-S, Yu J-T, Jiang T, et al. NLRP3 polymorphisms are associated with late-onset Alzheimer’s disease in Han Chinese. J Neuroimmunol. 2013;265(1–2):91–95. doi:10.1016/j.jneuroim.2013.10.002
25. Rheinheimer J, de Souza BM, Cardoso NS, Bauer AC, Crispim D. Current role of the NLRP3 inflammasome on obesity and insulin resistance: a systematic review. Metabolism. 2017;74:1–9. doi:10.1016/j.metabol.2017.06.002
26. Zhu Z, Yan J, Geng C, et al. A polymorphism Within the 3′ UTR of NLRP3 is associated with susceptibility for ischemic stroke in Chinese population. Cell Mol Neurobiol. 2016;36(6):981–988. doi:10.1007/s10571-015-0288-1
27. Lee YH, Bae SC. Association between functional NLRP3 polymorphisms and susceptibility to autoimmune and inflammatory diseases: a meta-analysis. Lupus. 2016;25(14):1558–1566. doi:10.1177/0961203316644336
28. Zhang Q, Fan H, Zhang J, Wang Y, Xing H. NLRP3 rs35829419 polymorphism is associated with increased susceptibility to multiple diseases in humans. Genet Mol Res. 2015;14(4):13968–13980. doi:10.4238/2015.October.29.17
29. Zhang M, Tan X, Huang J, et al. Association between two interleukin-2 gene polymorphisms and cancer susceptibility: a meta-analysis. Onco Targets Ther. 2016;9:2181–2192. doi:10.2147/OTT.S94761
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
