Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 11
Genetic polymorphism analysis of patients with primary hyperhidrosis
Authors Simes BC, Moore JP , Brown TC, Rushforth TJ, Bookout AL, Richardson CL
Received 20 June 2018
Accepted for publication 14 August 2018
Published 11 October 2018 Volume 2018:11 Pages 477—483
DOI https://doi.org/10.2147/CCID.S176842
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jeffrey Weinberg
Bryce C Simes,1 Joshua P Moore,1 Terry C Brown,1 Tyler J Rushforth,1 Angela L Bookout,2 Chante L Richardson1
1Alabama College of Osteopathic Medicine, Dothan, AL, USA; 2Southern Institute of Dermatology, Dothan, AL, USA
Background: Hyperhidrosis affects 220 million people worldwide. The hallmark of this condition is excessive sweating, which negatively impacts the social, emotional, and occupational lives of these individuals. A familial predisposition has been established; however, the specific genes involved have yet to be identified.
Objective: The aim of this study was to determine possible genetic variations contributing to primary hyperhidrosis, specifically single-nucleotide polymorphisms (SNPs).
Patients and methods: Twenty-one case and 21 control DNA samples were extracted and genotyped for 20 SNPs associated with the Butyrylcholinesterase (BCHE) and Cholinergic Receptor Nicotinic Alpha-7 subunit (CHRNA7) genes.
Results: For rs1126680, the –116A variant allele (P-value=0.15) was found only in hyperhidrosis patients who also had the K-variant allele (P-value=0.65) in rs1803274. Further analysis testing the null hypothesis of independence between the combined genotypes and case/control status yielded a P-value of 0.30.
Conclusion: Our results are consistent with previous research that shows the K-variant requires the –116A variant to be present in order to observe a decrease in BChE activity levels. These results are not statistically significant (P-value >0.05), but the exclusive association between the –116A and K-variants on the BCHE gene in hyperhidrosis patients warrants further investigation using a larger sample size.
Keywords: hyperhidrosis, butyrylcholinesterase, BCHE, cholinergic receptor nicotinic alpha-7 subunit, CHRNA7, K-variant, rs1803274, -116A variant, rs1126680, acetylcholinesterase
Introduction and background
Hyperhidrosis affects 220 million people worldwide. The hallmark of this condition is excessive sweating, which negatively impacts the social, emotional, and occupational lives of these individuals. Hyperhidrosis can either be classified as primary or secondary. In primary hyperhidrosis, there is no underlying medical condition which causes the patient’s excessive sweating. In secondary hyperhidrosis, medications or endocrine diseases are usually the actual cause of excessive sweating. The sweating is either localized or generalized and is experienced in the hands, axillae, feet, or face.1 Sweat is secreted from eccrine glands in response to cholinergic stimulation. These simple, coiled tubular glands assist in the maintenance of body temperature in response to exercise or heat exposure. There are between two and four million eccrine sweat glands mostly located on the hands, feet, head, axillae, and groin. Physiologic secretion of sweat is mediated primarily by the neurotransmitter acetylcholine (ACh).1
Current treatment options available for patients with hyperhidrosis have an anticholinergic mechanism of action. Over-the-counter aluminum-based antiperspirants are first-order therapeutics, whereas anticholinergic treatments are second-order therapeutics. Anticholinergics competitively inhibit ACh at muscarinic receptors. Glycopyrrolate (off-label) is one of the most commonly used drugs by physicians for hyperhidrosis. However, long-term usage and adverse side effects, such as dry mouth, confusion, dizziness, and blurred vision, could render this treatment option unsafe in some individuals.1 An additional option, especially for those with axillary hyperhidrosis, is an injection of botulinum toxin. Botulinum toxin treats hyperhidrosis by irreversibly blocking the release of ACh from presynaptic bulbs at postganglionic nerve endings.2 When all conservative treatment options fail, some patients opt to undergo an endoscopic thoracic sympathectomy. This method is relatively successful in resolving symptoms, but a significant portion of patients experience compensatory sweating.1
It is generally accepted that primary hyperhidrosis is a familial disorder. One study found that 62% of those who have primary hyperhidrosis reported a family history of the disease with an autosomal dominant pattern of inheritance.3 However, there is no exact known cause of hyperhidrosis, and thus, there has been minimal investigation into the specific genes involved.4 The aim of this study was to determine possible genetic variations contributing to primary hyperhidrosis, specifically single-nucleotide polymorphisms (SNPs). By linking a specific genetic cause to primary hyperhidrosis, the mechanism of action can be elucidated and assist in identification of new therapeutic targets.
Butyrylcholinesterase (BChE) and the BCHE gene
ACh is the neurotransmitter released by the preganglionic neurons of the sympathetic and parasympathetic nervous system as well as the postganglionic neurons of the parasympathetic nervous system. ACh is also the neurotransmitter released at the sweat glands of the sympathetic nervous system. Within the synaptic cleft, acetylcholinesterase (AChE) quickly hydrolyzes ACh into choline and acetate. Since AChE quickly hydrolyzes ACh, it is possible that AChE is involved in the regulation of sweating.5 BChE, also known as pseudocholinesterase, is synthesized in the liver and secreted into the blood. Until recently, the physiological role of BChE was unknown. BChE plays a supporting role in the hydrolysis of ACh that is apparent once AChE is inhibited.6 Another proposed role of BChE is that it functions like a bodyguard, protecting the ability of AChE to break down ACh in the synaptic cleft. This is because BChE has a much larger spectrum of compounds that it hydrolyzes when compared with AChE. In order for a cholinesterase inhibitor to act upon AChE, it theoretically may have to bypass BChE first.7
Studies have confirmed that genetic polymorphisms in the BCHE gene (Figure 1) influence the levels of serum-active AChE in the body, specifically rs2668207 and rs2048493. Howard et al suggest that “In participants with at least one minor allele, AChE levels were lower by 4.3%–9.5% at all time points” throughout the study.8 One SNP of interest on the BCHE gene is the K-variant (rs1803274), which replaces alanine with threonine at codon 539 (Figure 2).9 It is one of the most common and well-studied variations in the BCHE gene. It is associated with a 30% reduction in the activity of serum BChE due to the resulting structural instability.10 However, recent studies have found that also possessing the –116A (rs1126680) variant is necessary to observe the decrease in serum AChE levels.11,12
  | Figure 1 Idiogram of Chromosome 3 – BCHE gene on 3q26.1. Abbreviation: BCHE, Butyrylcholinesterase. |
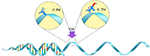  | Figure 2 BCHE K-variant – The alanine nucleotide is replaced by a threonine nucleotide at the 539 codon. Abbreviation: BCHE, Butyrylcholinesterase. |
The cholinergic receptor nicotinic Alpha-7 subunit and the CHRNA7 gene
The CHRNA 7 gene encodes the Cholinergic Receptor Nicotinic Alpha-7 subunit. Nicotinic ACh receptors are ligand-gated ion channels that mediate the effect of ACh and are highly expressed throughout the peripheral and central nervous systems. CHRNA 7 polymorphisms have been associated with cholinergic neurotransmission abnormalities observed in Alzheimer’s disease.13 Recent immunohistochemical research on patients with hyperhidrosis revealed that there is a higher expression of ACh and the cholinergic receptor nicotinic alpha-7 subunit in the sympathetic ganglion of those with hyperhidrosis compared with controls. The ganglion diameter was also larger in patients with hyperhidrosis.14
In summary, the physiology of sweating and effective treatment options available strongly suggest a cholinergic system pathophysiology in primary hyperhidrosis. The genetic evidence shows an autosomal dominant mode of inheritance, but research has yet to identify the specific genes involved. The purpose of this study was to determine genetic variations on the BCHE and CHRNA7 genes that may be responsible for primary hyperhidrosis.
Patients and methods
A non-invasive buccal swab was used to collect DNA samples from 21 case and 21 control participants. All participants completed an enrollment questionnaire containing the following elements: if they carry a diagnosis of hyperhidrosis, ingestion of glycoalkaloid plant foods, sweating severity on a scale of 1–10, farming occupation, history of delayed recovery after anesthesia and any other diagnosed endocrine, metabolic, or neurological medical conditions. Case participants were defined by either an official diagnosis of primary hyperhidrosis by a physician or self-reported by the patient. Control participants were defined as not having a diagnosis of primary hyperhidrosis and no subjective complaints of excessive sweating.
All samples were then sent to the University of Arizona Genetic Core for DNA extraction and genotyping. Twenty SNPs associated with the BCHE and CHRNA7 genes were chosen for analysis. For the BCHE gene: rs1126680, rs114706984, rs121918556, rs121918557, rs121918558, rs1355534, rs1355538, rs1799807, rs1803274, rs2048493, rs2668207, rs28933389, rs28933390, rs4263329, rs4680662, and rs829508. For the CHRNA7 gene: rs8024987, rs6494223, rs3087454, and rs1355920.
Genotyping was performed with Agena Mass Array for 19 of the 20 SNPs. The K-variant (rs1803274) on the BCHE gene was genotyped by Taqman assay to avoid proximal SNP errors. Testing for Hardy–Weinberg equilibrium, P-values were performed for each SNP using Graffelman’s exact test implemented into the Comprehensive R Archive Network software.15 P-values were calculated to test the null hypothesis of independence between variation and case/control status using the Fisher’s exact test algorithm.16 False Discovery Rate (FDR) P-values were also calculated to adjust for common errors seen in genetic association tests caused by multiple comparisons. The Benjamini–Hochberg procedure was chosen over the Bonferroni correction due to its statistical power advantage.17 This statistical analysis was further separated into cases with and without a diagnosis of primary hyperhidrosis from a physician.
Ethics approval
This study was approved by the Alabama College of Osteopathic Medicine Institutional Review Board on March 13, 2017.
Results
Sixteen SNPs on the BCHE gene and four on the CHRNA7 gene were successfully genotyped for all 42 participants in the two cohorts. A heatmap was generated to visualize this multivariate dataset, sorted by each SNP and sample number (Figure 3). The majority (88%) of the participants in both cohorts were of Caucasian descent. The average age for case participants was 47 years, and 37 years for control participants. A total of 71% of case participants were female compared with 52% of control participants. The body mass index (BMI) average for case participants was 29, and 27 for control participants. The average self-reported sweating severity in case participants was 7.7 out of 10 compared with 3.0 out of 10 for control participants. No significant differences in glycoalkaloid food consumption were observed between the case and control participants. Seven of the case participants did not carry an official diagnosis of primary hyperhidrosis from a physician but were still included due to the overall subjective nature of the diagnosis. Statistical analysis of all four SNPs on the CHRNA7 gene returned no significant evidence of association.
Of the forty-two participants, nine were heterozygous for the BCHE K-variant (rs1803274). A total of 67% of those with the K-variant were case participants and 33% control participants. One control participant (ACOM043B) was homozygous for the mutation but did not possess the –116A variant allele. All 21 control participants did not possess the –116A variant allele (Figures 4 and 5). For rs1126680, the –116A variant allele (P-value=0.15) was found only in hyperhidrosis patients who also had the K-variant allele (P-value=0.65) in rs1803274. The K-variant and the –116A variant are the two SNPs with the strongest observed association. When accounting for multiple comparisons, the FDR adjusted P-value, a measure of what percentage of statistically significant values will be false positives, for both SNPs was 1 (Table 1). Further analysis testing the null hypothesis of independence between the combined genotypes and case/control status yielded a P-value of 0.30.
In summary, 18 of the 20 SNPs that were investigated did not show an association with primary hyperhidrosis. However, the possession of both the –116A and K-variants on the BCHE gene was exclusively associated with cases of primary hyperhidrosis.
Discussion
The results of the study are consistent with previous research that shows the K-variant requires the –116A variant to be present in order to observe a decrease in BChE activity levels.12,19 Furtado et al found “Mean BChE activity in obese and control samples was significantly lower only when the −116A mutation was present” among all participant groups.11 Additional studies found that 50% of subjects with BChE activity <2000 U/L possessed the –116A variant.18
In our study, 15 out of 21 of the hyperhidrosis patients possessed neither the K-variant nor the –116A variant. Like many medical conditions, hyperhidrosis could be a multifactorial disease with genetic or lifestyle factors that the study did not account for. For example, sympathetic stimulants, such as caffeine could cause symptoms in patients with an otherwise normal cholinergic system. The subjective nature of diagnosis and lack of a definitive test for hyperhidrosis (ie, no blood test) may have also led to some cases having hyperhidrosis secondary to another medical condition, such as anxiety or hyperthyroidism.
The results of this study support the theory that the cholinergic system potentially plays a role in primary hyperhidrosis. The –116A and K-variants affect the structural stability of BChE, reducing its activity and ability to hydrolyze cholinesterase inhibitors. Future research confirming this association would allow for more advances in both the understanding and treatment options of primary hyperhidrosis.
Conclusion
In patients with primary hyperhidrosis, this study identified two SNPs of interest on the BCHE gene. These results are not statistically significant (P-value>0.05), but the exclusive association between the –116A and K-variants on the BCHE gene in hyperhidrosis patients warrants further investigation in future studies, including a larger sample size. The high Benjamini–Hochberg FDR-adjusted P-values observed in our study indicated a lack of statistical power that may be corrected by increasing the number of participants.
We suggest that future studies also investigate variables associated with the inhibition of AChE since the role of BChE was not evident until studies inhibited AChE. Some examples of variables include AChE inhibitors, such as organophosphates used in pesticides and natural glycoalkaloid toxins produced by plants of the Solanaceae (nightshade) family. These plants include commonly ingested foods, such as potatoes and tomatoes. Based on our hypothesis, those with variants in the BCHE gene would be more susceptible to inhibition of AChE, leading to hyperhidrosis symptoms.
While the SNPs that were studied for the CHRNA7 gene produced no statistically significant results, previous immunohistochemical studies showed overexpression of CHRNA7 receptors in the sympathetic ganglia of hyperhidrosis patients. Including a broader range of SNPs on the CHRNA7 gene in future studies could yield better results.
Acknowledgments
This study was supported and funded by the Southeast Alabama Medical Center Foundation, a 501(c)(3), not-for-profit public charity. Special thanks to the University of Arizona Genetics Core Laboratory and Dr Dennis Wylie of the University of Texas Center for Biomedical Research Support.
Consent for publication
Signed informed consent for our study was received from all 42 patients and will be kept at least 3 years beyond the study’s completion.
Disclosure
The authors report no conflicts of interest in this work.
References
Sito G. Hyperhidrosis: Clinicians Guide to Diagnosis and Treatment. Cham: Springer; 2016. | ||
Singh S, Davis H, Wilson P. Axillary hyperhidrosis: a review of the extent of the problem and treatment modalities. Surgeon. 2015;13(5):279–285. | ||
Kaufmann H, Saadia D, Polin C, Hague S, Singleton A, Singleton A. Primary hyperhidrosis—evidence for autosomal dominant inheritance. Clin Auton Res. 2003;13(2):96–98. | ||
Shargall Y, Spratt E, Zeldin RA. Hyperhidrosis: what is it and why does it occur? Thorac Surg Clin. 2008;18(2):125–132. | ||
Shibasaki M, Crandall CG. Effect of local acetylcholinesterase inhibition on sweat rate in humans. J Appl Physiol. 2001;90(3):757–762. | ||
Lockridge O, Duysen EG, Masson P. Butyrylcholinesterase: overview, structure, and function. Anticholinesterase Pesticides. 2011:25–41. | ||
Giacobini E. Cholinesterases and Cholinesterase Inhibitors. London: Martin Dunitz; 2001. | ||
Howard TD, Hsu FC, Grzywacz JG, et al. Evaluation of candidate genes for cholinesterase activity in farmworkers exposed to organophosphorus pesticides: association of single nucleotide polymorphisms in BCHE. Environ Health Perspect. 2010;118(10):1395–1399. | ||
Pongthanaracht N, Yanarojana S, Pinthong D, et al. Association between butyrylcholinesterase K variant and mild cognitive impairment in the Thai community-dwelling patients. Clin Interv Aging. 2017;12:897–901. | ||
Wang Z, Jiang Y, Wang X, et al. Butyrylcholinesterase K variant and Alzheimer’s disease risk: a meta-analysis. Med Sci Monit. 2015;21:1408–1413. | ||
Furtado-Alle L, Andrade FA, Nunes K, Mikami LR, Souza RL, Chautard-Freire-Maia EA. Association of variants of the -116 site of the butyrylcholinesterase BCHE gene to enzyme activity and body mass index. Chem Biol Interact. 2008;175(1–3):115–118. | ||
Chaves TJ, Leite N, Milano GE, et al. −116A and K BCHE gene variants associated with obesity and hypertriglyceridemia in adolescents from Southern Brazil. Chem Biol Interact. 2013;203(1):341–343. | ||
Weng PH, Chen JH, Chen TF, et al. CHRNA7 polymorphisms and response to cholinesterase inhibitors in Alzheimer’s disease. PLoS One. 2013;8(12):e84059. | ||
de Moura Júnior NB, das-Neves-Pereira JC, de Oliveira FR, et al. Expression of acetylcholine and its receptor in human sympathetic ganglia in primary hyperhidrosis. Ann Thorac Surg. 2013;95(2):465–470. | ||
Graffelman J. Exploring diallelic genetic markers: the HardyWeinberg package. J Stat Softw. 2015;64(3). | ||
Mehta CR, Patel NR. ALGORITHM 643: FEXACT: a FORTRAN subroutine for Fisher’s exact test on unordered r×c contingency tables. ACM Trans Math Softw. 1986;12(2):154–161. | ||
Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Statist. 2001;29(4):1165–1188. | ||
Jasiecki J, Jońca J, Żuk M, et al. Activity and polymorphisms of butyrylcholinesterase in a Polish population. Chem Biol Interact. 2016;259(Pt B):70–77. | ||
Lima JK, Leite N, Turek LV, et al. 1914G variant of BCHE gene associated with enzyme activity, obesity and triglyceride levels. Gene. 2013;532(1):24–26. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




