Back to Journals » Infection and Drug Resistance » Volume 13
Genetic Diversity of Schistosoma haematobium in Qena Governorate, Upper Egypt
Authors El-Kady AM , EL-Amir MI , Hassan MH , Allemailem KS , Almatroudi A , Ahmad AA
Received 9 June 2020
Accepted for publication 15 September 2020
Published 15 October 2020 Volume 2020:13 Pages 3601—3611
DOI https://doi.org/10.2147/IDR.S266928
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Asmaa M El-Kady,1 Mostafa I EL-Amir,2 Mohammed H Hassan,3 Khaled S Allemailem,4 Ahmad Almatroudi,4 Alzahraa Abdelraouf Ahmad5
1Department of Medical Parasitology, Faculty of Medicine, South Valley University, Qena, Egypt; 2Department of Medical Microbiology and Immunology, Faculty of Medicine, South Valley University, Qena, Egypt; 3Department of Medical Biochemistry, Faculty of Medicine, South Valley University, Qena, Egypt; 4Department of Medical Laboratories, College of Applied Medical Sciences, Qassim University, Buraydah, Saudia Arabia; 5Department of Medical Parasitology, Faculty of Medicine, Assiut University, Assiut, Egypt
Correspondence: Asmaa M El-Kady
Department of Medical Parasitology, Faculty of Medicine, South Valley University, Qena, Egypt
Tel +201114229741
Email [email protected]
Introduction: Schistosomiasis is an important neglected tropical disease (NTD) in several developing countries. Praziquantel is the principle and efficacious chemotherapeutic agent that has been used to treat schistosomiasis for decades. Unfortunately, emerging resistance to praziquantel with accompanying reduced efficacy is reported in some localities. Hence, genetic diversity among parasite populations is of significant interest in assessing the effects of selective pressure generated by praziquantel therapy that might result in encouraging the emergence of new genotypes that are either non-susceptible or drug-resistant. The present study aimed to investigate the genetic diversity of Schistosoma haematobium among human populations using the RAPD technique to help clarify disease epidemiology and transmission.
Materials and Methods: S. haematobium eggs were isolated from 50 of 134 patients from four different localities in Qena Governorate, Upper Egypt. These patients complained of terminal hematuria and burning micturition. Samples were used for molecular analysis using RAPD-PCR primers (A02, A07, A09, A10).
Results: Twenty S. haematobium isolates (40%) were amplified using the selected RAPD primers. Amplification patterns of these isolates showed distinct variation in the size and number of amplified fragments, indicating high genetic variation among these isolates.
Conclusion: To the best of our knowledge, this study is the first to characterize the genetic diversity of S. haematobium in human populations in Upper Egypt. Future studies on a larger geographic scale involving many districts in Upper Egypt should be encouraged. Information from such a study would provide better insight into clonal lineages of S. haematobium in this endemic area. In turn, understanding transmission of the parasite may have a major role in establishing control strategies for urogenital schistosomiasis in Upper Egypt.
Keywords: Schistosomiasis haematobium, RAPD, Upper Egypt, genetic diversity
Introduction
Schistosomiasis is an important neglected tropical disease (NTD). The responsible parasite is considered second only to malaria for its socioeconomic impacts.1 Worldwide, approximately 779 million people are at risk of infection in endemic areas and more than 250 million people are estimated to be currently infected with Schistosomiasis haematobium.2 The majority of recorded cases are concentrated in Africa, account for nearly 90% of infections.3–5 In developing countries, schistosomiasis causes disabilities in over 3.3 million people and 11, 700 deaths annually. The parasite remains a global threat even in non-endemic areas.6,7 Since ancient times, urogenital and intestinal schistosomiasis have been prevalent conditions in Egypt.8
Six species of schistosomes that infect humans are endemic in 78 countries, namely; Schistosoma mansoni, S. haematobium, S. japonicum, S. intercalatum, S. mekongi, and S. guineensis.9 S. haematobium causes urogenital schistosomiasis and shows an important association with viral infections, such as HIV and HPV.10 The species occurs in a broad geographical range across Africa, parts of the Middle East, Madagascar and the Indian Ocean Islands. Various Bulinid snails serve as intermediate hosts.11 The estimated prevalence rate of human infections caused by S. haematobium exceeds 112 million cases in sub-Saharan Africa, more infections than estimated for other schistosome species.1 In Egypt, the prevalence of S. haematobium has decreased, now ranging from 0% to 13.9%, due to the implementation of national control programs.12,13
The clinical picture of S. haematobium varies widely, ranging from mild hematuria with or without gross bladder wall pathology to serious renal disease. Cancer of the bladder may be associated with the course of the disease. These variations in clinical presentation of the disease may change according to screening policies and delays in diagnosis.1,14
In several sub-Saharan countries, national control programs are implemented to reduce the burden of schistosomiasis through mass administration of anthelmintic drugs, particularly regions with a high endemic incidence of infection.15,16 Praziquantel is the principal agent for treatment of schistosomiasis; the drug has been used for this purpose for decades.17 Mass drug administration may, however, influence (if not correctly done) the prevalence of S. haematobium in migrants to low-endemic countries.18 Moreover, emerging resistance with reduced efficacy is reported for praziquantel against human schistosomiasis in some localities.19,20 Several clinical trials conducted in Senegal and Egypt show reduced effectiveness of praziquantel against schistosomes and isolated drug-resistant strains.21–23 A previous study conducted of Egyptian villagers reported inability of PZQ in curing about 1–2.4% of 1000 schistosomiasis-infected patients. They also detected PZQ-resistant strains that can withstand high doses of the drug.24
Recent molecular epidemiological studies of schistosomiasis provide knowledge of the dynamics of parasitic infection, understanding of genetic characteristics of control measures, and models for evolution and the spread of drug resistance.22,23,25 Genetic differences among schistosomes can lead to the emergence of drug-resistant strains and possibly create non-susceptible genotypes.27,28
Yet, genetic variation in S. haematobium remains understudied relative to S. mansoni, mainly as a result of the extensive requirements for its laboratory maintenance and a deficiency in specific molecular markers.29,30
Recent studies have used randomly amplified polymorphic DNA (RAPD)-PCR as a valuable method to explore the genetic diversity of schistosome populations in snails despite assumptions required for its use.31–33 RAPD primers screen a wide range of loci across the entire schistosomal genome with limited available sequence data and low DNA yield. The technique makes it more feasible and proved to be effective in characterizing inter-and intra-specific relationships.34,35
In the present study, the diversity of S. haematobium among infected patients from Qena Governorate, Upper Egypt, was explored using the RAPD technique to clarify the molecular structure of natural schistosome populations and understand aspects of disease epidemiology and transmission. This study is the first, to the best of our knowledge, to examine the genetic diversity of S. haematobium in human populations in Upper Egypt.
Materials and Methods
Study Location
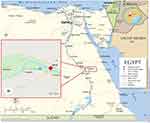
The present study was conducted in Qena Governorate, Upper Egypt, approximately 608 km south of Cairo. The Governorate extends for 240 km (Figure 1) and is connected to the north by Sohag, the south by Aswan, the east by the Red Sea, and the west by the New valley. The population of the Governorate was 2.8 million in 2004.
Sampling and Collection of S. haematobium Eggs
Sampling was conducted in Qena General Hospital over the period from October 2018 to February 2019. One hundred thirty-four patients were recruited from the urology outpatient clinic, all with suspected urinary schistosomiasis. Subjects complained of terminal hematuria and burning micturition. Participants provided a history of contact with water either by irrigation, swimming or washing utensils. Participants were from four different localities in Qena Governorate (Figure 1 and Table 1). Three rural areas; Al hujyrat, Al tramsa, and Dandara, and one urban locality (Qena city) were represented. The age range of patients enrolled in the study was 1 to 65 years. Most subjects were male (82, 61.2%).
 |
Table 1 No. of the Included Participants in the Study and Their Distribution |
Parasitological Examination
A 10 mL sample of urine was collected from each participant in a clean dry container. Personal data of each participant (name, sex, age, and residence) was recorded, and urine samples were processed within 1–3 hours of collection. Each sample was concentrated by simple sedimentation and examined for S. haematobium eggs in the parasitology laboratory. Microscopically positive samples were preserved at −20 °C until molecular analysis.
Molecular Analysis
DNA from positive samples extracted using a GeneJET Whole Blood Genomic DNA Purification Mini Kit (Thermo Scientific, Cat. No. K0781). The Concentration and purity of DNA were estimated. Four 10-bp oligonucleotide primers (Operon Technologies, Alameda, CA, USA) were used: Primer A02: TGCCGAGCTG, A07: GAAACGGGTG, A09: GGGTAACGCC, and A10: GTGATCGCAG. Isolated DNA of S. haematobium was used for PCR according to Shiff et al36 in a thermal cycler (Veriti ™ 96-well thermal cycler (9902, Singapore). The reaction volume was 20 µL, and PCR conditions were adjusted to 40 cycles of denaturation for 10s at 94 ◦C, annealing for 1 min at 36 ◦C, and extension for 2 minutes at 72 ◦C). Samples separated together with a 100 bp DNA Ladder (Thermo Fisher Scientific, Waltham, Massachusetts, USA) in 1.2% agarose gel electrophoresis stained with ethidium bromide. The PCR products were then visualized and photographed under UV light. Images were analyzed with a Gel Imager and Documentation System (Compact M, Biometra, Germany).
Data Analysis
The amplified RAPD-PCR products produced for each human isolate were compared to the DNA ladder for proper band score and to determine the molecular weights of DNA fragments. All the amplified bands produced by each primer in separate patient isolates have been pooled. A comparison between isolates was done by comparing total bands and shared bands among isolates. Genetic distances among isolates were measured using RAPD Distance 1.04 software.37 A binary matrix was constructed on a pair-wise basis, based assigning presence or absence of RAPD bands as either 1 or 0.38,39 The similarity coefficients between isolates were estimated by simple association, and Nei and Li.40 A dendrogram was developed using UPGMA (unweighted pair group method with arithmetic mean) constructed by SPSS version 16 (SPSS Inc., Chicago, IL, United States).
Results
Demographic Data of the Patients
One hundred and thirty-four patients participated in the study. All subjects complained of symptoms suggestive of urinary schistosomiasis (hematuria and dysuria) and visited the urology outpatient clinic in Qena General Hospital for diagnosis and treatment. Most patients were male (61.2%), mainly from rural areas (60.4%). Hematuria was the main symptomatology (88.1%), while dysuria was the complaint in only 3% of cases. A combination of hematuria and dysuria presented in 9% of patients. Urine examination revealed 50/134 patients (37.3%) were positive for schistosomiasis, S. haematobium. Seventy percent of positive cases were in males, and 84% complained of hematuria.
Molecular Characterization of S. haematobium
Fifty microscopically positive S. haematobium urine samples were used for RAPD-PCR. The extracted DNA showed variable DNA yields from low to very high concentrations (7.4–709.5 ng/µL.). DNA from 20 samples (40%) was amplified by the selected RAPD primers (A02, A07, A09, A10). However, a distinct variation in amplification banding pattern was observed with the 20 positive results. Variation in the size and number of amplified fragments from each primer in each isolate was also seen. Amplified fragments showed differences in intensity under UV light. Criteria for patients positive by RAPD–PCR are shown in Table 2.
 |
Table 2 Criteria of Patients Gave Positive RAPD –PCR Results |
RAPD Polymorphism Among Schistosoma haematobium Human Isolates
The results of this study showed that the four RAPD primers amplified 40 distinct DNA fragments with an average number of bands per primer ranging from nine to 12. The size of the amplified fragments ranged from 198 bp with A09 primer to 1860 bp with A10 primer. All DNA fragments were polymorphic (Figure 2, Table 3).
 |
Table 3 RAPD Polymorphism Among Human Schistosoma haematobium Isolates |
Out of a total of 40 generated bands, a high similarity was observed between 23 bands of the DNA amplified from the 20 human isolates of S. haematobium. Primer A02 generated PCR products for 20 human samples while primers (A07, A09, and A10) amplified 18 human isolates. Human isolates (H) nos. 44 and 45 were amplified only by primer A02. Regarding Primer A02, many shared bands were observed among human isolates at the following sites; 470 bp, 604 bp, and 794 bp. However, the polymorphic band at 698 bp was amplified by only one human isolate (H35) with PIC (polymorphic information content) of about 0.29. A high degree of polymorphism was also seen with primer A07 among most isolates. Genomic DNA of human isolates showed similarity at 604 bp and 987 bp, while bands at 880 bp, 195 bp, and 1171 bp were least shared. Primer A07 showed the highest PIC (0.41).
Primer A09 showed the highest number of polymorphic bands (12). DNA fragments at 527 bp and 902 bp were amplified in almost all samples with high similarity. In contrast, DNA fragments at 1316 bp, 1527 bp, and 222 bp were the least shared among human isolates with PIC of 0.29. Conversely, primer A10 showed the lowest number of shared DNA fragments, especially at 241bp, 469 bp, and 1072 bp. The DNA band at 1325 bp was amplified by nearly all samples and PIC, 0.27, was the smallest among primers (0.27).
Similarity coefficients for four RAPD markers among the 20 human isolates ranged from 0.00 to 0.95 (Table 4). The greatest similarity was found between human isolates H9 and H15 (95%), followed by H3 and H47 (88.9%). Notably, human isolate H44 showed 100% polymorphism among all isolates. The constructed dendrogram using UPGMA clustering method, based on Nei and Li coefficients, was generated from all amplified bands obtained by the four RAPD primers (Figure 3). The analysis shows 40 genetic loci produced by the four RAPD primers grouped into five main clusters and 18 groups. A close relation between isolates of S. haematobium is observed. Cluster (F2) comprised four sub-clusters containing 13 isolates. The first sub-cluster is comprised of human isolates 47, 36, 34, and 30; the second of 31 and 13 with a 0.815 similarity coefficient. The third sub-cluster consists of isolates 39, 21, 11, 15, and 9, and the fourth sub-cluster, 37 and 7 with a 0.813 similarity coefficient. Human isolates 44, 45, 42, 35, and 38 formed five separate and distinct lines A1, B1, C1, E1, and F1, respectively, showing dissimilarity with other S. haematobium isolates.
 |
Table 4 Genetic Similarity Coefficient Among Human S. haematobium Isolates Showing Dissimilarities Between Isolates |
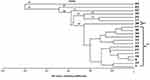 |
Figure 3 Dendrogram (UPGMA) based on Nei and Li similarity coefficient. |
Discussion
The present study shows that the prevalence of schistosomiasis caused by S. haematobium in patients with urologic manifestations at the urologic outpatient clinic at Qena General Hospital, was 37.3%; most patients were males from rural areas. Such a prevalence rate is higher than previously documented, which might be explained by small sample size and the selective sampling of patients complaining of hematuria and dysuria. Also, it could be attributed to the agricultural nature of Qena governorate; the main occupation in the region is farming. In any case, previous studies reported a lower prevalence ranging from 7.04% to 20.7%.13,41,42
The national schistosomiasis control program NSCP implemented a strategy for the elimination of schistosomiasis caused by S. haematobium in Egypt, based on interruption of its transmission by its intermediate host and mass therapy with praziquantel.12 Despite the consistent reduction in the prevalence of infection, some governorates in Upper Egypt, Sohag, Qena, and Aswan, still show a significant incidence of disease.43
Praziquantel has demonstrated high efficacy in disease control in various endemic regions, although its use has led to the emergence of strains that tolerate large doses of the medication through its widespread use in mass control measures.24
Many studies have discussed the genetic diversity among schistosome populations, whether S. haematobium or S. mansoni. Genetic diversity can be affected by various factors, such as overlapping contact sites, immunity, and susceptibility of the final and snail intermediate host. Further, human movements can promote co-existence of variable genotypes in certain regions. High genetic diversity may also contribute to the emergence of new strains that are either non-susceptible or drug-resistant through genetic swapping and recombination between old and newly emerged genotypes.27,44
This study was designed to detect genetic diversity among S. haematobium populations in infected patients in Qena governorate, Upper Egypt using RAPD-PCR. Results help clarify the epidemiology of schistosomiasis, and confirm the persistence of S. haematobium in Upper Egypt despite national control programs.
Randomly amplified polymorphic DNA markers represent a valuable method for evaluating the genetic diversity and gene flow among parasitic populations in humans and their intermediate hosts. A study performed by Dabo et al used RAPD primers to detect the presence of multiple genotypes of S. haematobium within its intermediate host snails.45 A previous study in Egypt used RAPD-PCR analysis for natural S. haematobium populations to analyze the genetic diversity of Egyptian isolates as opposed to other African isolates.46
In the present research, four RAPD markers A02, A07, A09, and A10 were tested against genomic DNA isolated from 50 S. haematobium egg samples. These primers proved to be useful in differentiating S. haematobium from other schistosomes. However, the present RAPD banding pattern gave positive results with only 20 samples, indicating that the number of S. haematobium eggs was not sufficient for genomic DNA isolation. Moreover, DNA yields varied greatly, from 7.4 to 709.5 ng/µL. Differences in the amounts of DNA might affect PCR results, especially in the absence of pooled urine samples technique that could increase the possibility of detection of S. haematobium infection.47 Some studies discuss the efficacy of DNA pooling in evaluating genetic diversity of a given population, from which allele frequencies could be calculated.48 However, several studies also discuss controversial results of using pooled DNA samples to assess genetic variation.48–50
Data present here show distinct polymorphic patterns from DNA amplified with A02, A07, A09, and A10 primers. These primers are the same as the primers endorsed by Afifi et al. These authors used the same primers to amplify genomic DNA isolated from S. haematobium eggs found in the urine of infected patients. Findings exhibited moderate to high variation in genomic schistosome populations, thus clarifying genetic diversity among S. haematobium populations in Egypt relative to other African isolates from Zimbabwe and South Africa.46 Our results exhibited high genetic diversity among human isolates in four neighboring geographical regions in Qena Governorate, Upper Egypt. These results are in harmony with those of previous studies that document a high degree of genetic variation in selected S. haematobium populations.51,52
Results of the current study indicate that estimated genetic similarity denoted by Nei and Li (1979) coefficients of similarity using numbers of shared bands across isolates varied greatly, from 0% to 95%. These findings suggest significant intraspecies differences among human isolates included in the study.40 Even though 100% polymorphism was detected, some human isolates (H9 and H15, and H36 and H47) were closely related and showed high similarity percentages of 95% and 88.9%, respectively. Moreover, results of UPGMA analysis showed that the four RAPD primers generated five main clusters and 18 groups within S. haematobium isolates. Human isolates, 44, 45, 42, 35, and 38, formed separate and distinct lines A1, B1, C1, E1, and F1, respectively, showing dissimilarity with the other 16 S. haematobium isolates. The presence of hybrids in those populations could provide an explanation for this finding. Hybridization and introgression with other Schistosoma species are reported in other parts of Africa.53–55 However, hybridization could not be confirmed in the present study in the absence of comparative control isolates of different schisosomes species..
The high percentage of intraspecies variability among the studied S. haematobium populations could be due to variations in DNA quality, contamination, and instrumentation, which can generate substantial sample-to-sample deviation.56 Conversely, it could be attributed to the high prevalence rate in the Qena region. Our findings are consistent with a previous study that shows a high allelic variation of S. haematobium found in human hosts in Mali and Nigeria. We hypothesize that these findings may result from a higher prevalence of parasitic infection in those localities.50 Unlike our findings, other studies showed unexpectedly low genetic diversity in S. haematobium populations using DNA barcoding methods.57
Many other parts in the world show high diversity in Schistosoma populations, such as Yemen Zanzibar, the neighboring African coastal region, and the Indian Islands.27,58 The level of genetic diversity is a significant measure for assessing the effects of selective pressure generated by drug therapy control measures. Hence, high genetic diversity may provide a sizable genetic basis for selection to increase the rate of development of praziquantel resistance. Eventually, the effect of mass praziquantel administration in control programs on the genetic selection in Schistosoma populations could promote a decline in genetic diversity and the evolution of new non-susceptible or resistant strains.52 Alterations in parasite diversity associated with drug therapy raise the need for proper monitoring in endemic areas.
Conclusion
To our knowledge, this study is the first to characterize the genetic diversity of S. haematobium in human populations in Upper Egypt using RAPD markers. It provides new insights into the population genetics of S. haematobium in this endemic area. RAPD primers used proved to be valuable and cost-effective tools for assessing the genetic diversity of S. haematobium. Future studies on a larger geographic scale involving many districts in Upper Egypt should be encouraged to screen more parasite isolates with different primers and to investigate its intermediate host. Information from such studies would provide better insight into the clonal lineages of S. haematobium. This knowledge might play a major role in establishing control strategies for urogenital schistosomiasis in Upper Egypt.
Ethical Statement
All procedures performed in the present study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. This research had an ethical approval by the institutional review board of the faculty of medicine, South Valley University, Qena, Egypt. Informed consent from all adult patients and the guardians of children participating in the study were obtained.
Acknowledgments
The authors acknowledge Dr. Ameer Elfarash, Department of Genetics, Faculty of Agriculture, Assiut University for his valuable effort in data analysis of RAPD technique.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by a grant from south valley university, Qena, Egypt (Grant No.4/2018). The funders had no role in study design, data collection, analysis or interpretation of the data or preparation of the manuscript.
Disclosure
We (the authors) declare that we have no conflicts of interest for this work.
References
1. Van der Werf MJ, De Vlas SJ, Brooker S, et al. Quantification of clinical morbidity associated with schistosome infection in sub-Saharan Africa. Acta Trop. 2003;86(2–3):125–139.
2. Molyneux DH, Savioli L, Engels D. Neglected tropical diseases: progress towards addressing the chronic pandemic. Lancet. 2017;389(10066):312–325. doi:10.1016/S0140-6736(16)30171-4
3. Hotez PJ, Alvarado M, Basáñez MG, et al. The global burden of disease study 2010: interpretation and implications for the neglected tropical diseases. PLoS Negl Trop Dis. 2014;8(7):e2865. doi:10.1371/journal.pntd.0002865
4. Colley DG, Secor WE. Immunology of human schistosomiasis. Parasite Immunol. 2014;36(8):347–357. doi:10.1111/pim.12087
5. Hay SI, Abajobir AA, Abate KH, et al. Global, regional, and national disability-adjusted life-years (DALYs) for 333 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2016: a systematic analysis for the global burden of disease study 2016. Lancet. 2017;390(10100):1260–1344.
6. Comelli A, Riccardi N, Canetti D, et al. Delay in schistosomiasis diagnosis and treatment: a multicenter cohort study in Italy. J Travel Med. 2020;27(1):taz075. doi:10.1093/jtm/taz075
7. Mantica G, Van der Merwe A, Terrone C, et al. Awareness of European practitioners toward uncommon tropical diseases: are we prepared to deal with mass migration? Results of an international survey. World J Urol. 2020;38(7):1773–1786. doi:10.1007/s00345-019-02957-73
8. Haggag AA, Rabiee A, Abd Elaziz KM, Gabrielli AF, Hay RA, Ramzy RMR. Mapping of schistosoma mansoni in the Nile Delta, Egypt: assessment of the prevalence by the circulating cathodic antigen urine assay. Acta Trop. 2020;6(167):9–17.
9. Colley DG, Bustinduy AL, Secor WE, King CH. Human schistosomiasis. Lancet. 2014:2253–2264
10. Bustinduy A, King C, Scott J, et al. HIV and schistosomiasis co-infection in African children. Lancet Infect Dis. 2014;14(7):640–649. doi:10.1016/S1473-3099(14)70001-5
11. Rollinson D, Stothard J, Southgate V. Interactions between intermediate snail hosts of the genus Bulinus and schistosomes of the Schistosoma haematobium group. Parasitology. 2001;123(Suppl. 7):45–60. doi:10.1017/S0031182001008046
12. Haggag AA, Rabiee A, Abd Elaziz KM, et al. Elimination of schistosomiasis haematobia as a public health problem in five governorates in Upper Egypt. Acta Trop. 2018;188:9–15. doi:10.1016/j.actatropica.2018.08.024
13. Sayed FG, Alsalahy M, Abd-El-Kader MS. Prevalence of schistosomiasis among patients attending Qena general hospital over the years 2011 and 2012. J Egypt Soc Parasitol. 2014;44(2):489–495. doi:10.12816/0006487
14. Joubert PH, Pretorius SJ, Kruger FJ. Further studies on the susceptibility of Bulinus africanus to infection with Schistosoma haematobium. Ann Trop Med Parasitol. 1991;85(2):253–258. doi:10.1080/00034983.1991.11812553
15. Stothard JR, Chitsulo L, Kristensen TK, Utzinger J. Control of schistosomiasis in sub-Saharan Africa: progress made, new opportunities and remaining challenges. Parasitology. 2009;136(13):1665–1675. doi:10.1017/S0031182009991272
16. Savioli L, Gabrielli AF, Montresor A, Chitsulo L, Engels D. Schistosomiasis control in Africa: 8 years after world health assembly resolution 5419. Parasitology. 2009;136(13):1677–1681. doi:10.1017/S0031182009991181
17. King CH, Mahmoud AAF. Drugs five years later: praziquantel. Ann Intern Med. 1989;110(4):290–296. doi:10.7326/0003-4819-110-4-290
18. Riccardi N, Nosenzo F, Peraldo F, et al. Increasing prevalence of genitourinary schistosomiasis in Europe in the migrant era: neglected no more? PLoS Negl Trop Dis. 2017;11(3):e0005237. doi:10.1371/journal.pntd.0005237
19. Davis A, Wegner DHG. Multicentre trials of praziquantel in human schistosomiasis: design and techniques. Bull World Health Organ. 1979;57(5):767.
20. Brindley PJ. Drug resistance to schistosomicides and other anthelmintics of medical significance. Acta Trop. 1994;56(2–3):213–231. doi:10.1016/0001-706X(94)90064-7
21. Picquet M, Vercruysse J, Shaw DJ, Diop M, Ly A. Efficacy of praziquantel against Schistosoma mansoni in northern Senegal. Trans R Soc Trop Med Hyg. 1998;92(1):90–93. doi:10.1016/S0035-9203(98)90971-3
22. Guisse F, Polman K, Stelma FF, et al. Therapeutic evaluation of two different dose regimens of praziquantel in a recent Schistosoma mansoni focus in northern Senegal. Am J Trop Med Hyg. 1997;56(5):511–514. doi:10.4269/ajtmh.1997.56.511
23. Bennett JL, Day T, Feng-Tao L, Ismail M, Farghaly A. The development of resistance to anthelmintics: a perspective with an emphasis on the antischistosomal drug praziquantel. Exp Parasitol. 1997;87(3):260–267. doi:10.1006/expr.1997.4229
24. Ismail M, Metwally A, Farghaly A, Bruce J, Tao LF, Bennett JL. Characterization of isolates of Schistosoma mansoni from Egyptian villagers that tolerate high doses of praziquantel. Am J Trop Med Hyg. 1996;55(2):214–218. doi:10.4269/ajtmh.1996.55.214
25. Norton AJ, Gower CM, Lamberton PHL, et al. Genetic consequences of mass human chemotherapy for Schistosoma mansoni: population structure pre-and post-praziquantel treatment in Tanzania. Am J Trop Med Hyg. 2010;83(4):951–957. doi:10.4269/ajtmh.2010.10-0283
26. GowerCM, GabrielliAF, SackoM, DembeleR, GolanR, EmeryAMRollinsonD, WebsterJP.Population genetics of Schistosoma haematobium: development of novel microsatellite markers and their application to schistosomiasis control in Mali. Parasitology. 2011;138:978–994
27. Sady H, Al-Mekhlafi HM, Webster BL, et al. New insights into the genetic diversity of Schistosoma mansoni and S. haematobiumin Yemen. Parasit Vectors. 2015;8(1):1–14. doi:http://dx.doi.10.1186/s13071-015-1168-8
28. Brouwer KC, Ndhlovu PD, Wagatsuma Y, Munatsi A, Shiff CJ. Urinary tract pathology attributed to Schistosoma haematobium: does parasite genetics play a role? Am J Trop Med Hyg. 2003;68(4):456–462. doi:10.4269/ajtmh.2003.68.456
29. Van den Broeck F, Geldof S, Polman K, Volckaert FAM, Huyse T. Optimal sample storage and extraction protocols for reliable multilocus genotyping of the human parasite Schistosoma mansoni. Infect Genet Evol. 2011;11(6):1413–1418. doi:10.1016/j.meegid.2011.05.006
30. Golan R, Gower CM, Emery AM, Rollinson D, Webster JP. Isolation and characterization of the first polymorphic microsatellite markers for Schistosoma haematobium and their application in multiplex reactions of larval stages. Mol Ecol Resour. 2008;8(3):647–649. doi:10.1111/j.1471-8286.2007.02031.x
31. Minchella DJ, Lewis FA, Sollenberger KM, Williams JA. Genetic diversity of Schistosoma mansoni: quantifying strain heterogeneity using a polymorphic DNA element. Mol Biochem Parasitol. 1994;68(2):307–313. doi:10.1016/0166-6851(94)90175-9
32. Sire C, Durand P, Pointier J-P TA. Genetic diversity and recruitment pattern of schistosoma mansoni in a biomphalaria glabrata snail population: a field study using random-amplified polymorphic DNA markers. J Parasitol. 1999;85(3):436. doi:10.2307/3285775
33. Lynch M, Milligan BG. Analysis of population genetic structure with RAPD markers. Mol Ecol. 1994;3(2):91–99. doi:10.1111/j.1365-294X.1994.tb00109.x
34. Barra V, This P, Imbert-Establet D, Combes C, Delseny M. Genetic variability and evolution of the Schistosoma genome analysed by using random amplified polymorphic DNA markers. Mol Biochem Parasitol. 1993;59(2):211–221. doi:10.1016/0166-6851(93)90219-N
35. Dias Neto E, Pereira de Souza C, Rollinson D, Katz N, Pena SDJ, Simpson AJG. The random amplification of polymorphic DNA allows the identification of strains and species of schistosome. Mol Biochem Parasitol. 1993;57(1):83–88. doi:10.1016/0166-6851(93)90246-T
36. Shiff C, Brouwer KC, Clow L. Schistosoma haematobium: population genetics of S. haematobium by direct measurement of parasite diversity using RAPD ± PCR. Exp Parasitol. 2000;96(1):47–51. doi:10.1006/expr.2000.4548
37. Armstrong J, Gibbs A, Peakall R. The RAPDistance Package. 1994.
38. Mattner J, Zawko G, Rossetto M, Krauss SL, Dixon KW, Sivasithamparam K. Conservation genetics and implications for restoration of Hemigenia exilis (lamiaceae), a serpentine endemic from Western Australia. Biol Conserv. 2002;107(1):37–45. doi:10.1016/S0006-3207(02)00041-1
39. Mekuria GT, Collins G, Sedgley M. Genetic diversity within an isolated olive (Olea europaea L.) population in relation to feral spread. Sci Hortic (Amsterdam). 2002;94(1–2):91–105. doi:10.1016/S0304-4238(01)00375-2
40. Nei M, Li W-H. Mathematical model for studying genetic variation in terms of restriction endonucleases (molecular evolution/mitochondrial DNA/nucleotide diversity). Genetics. 1979;76(10):5269–5273.
41. El-Khoby T, Galal N, Fenwick A, et al. The epidemiology of schistosomiasis in Egypt: summary findings in nine governorates. Am J Trop Med Hyg. 2000;62(2SUPPL):88–99. doi:10.4269/ajtmh.2000.62.88
42. Hussein AA, Bin-dajem SM. Prevalence of urinary schistosomiasis and infections with trematode larval stages in Bulinus truncatus snails from Qena Upper Egypt. J Appl Sci Res. 2008;4(11):1610–1617.
43. Barakat RMR. Epidemiology of schistosomiasis in Egypt: travel through time: review. J Adv Res. 2020;4:425–432. doi:http://dx.doi.10.1016/j.jare.2012.07.003
44. Brouwer KC, Ndhlovu P, Munatsi A, Shiff CJ. Genetic diversity of a population of Schistosoma haematobium derived from schoolchildren in East Central Zimbabwe. J Parasitol. 2001;87(4):762. doi:10.1645/0022-3395(2001)087[0762:GDOAPO]2.0.CO;2
45. Dabo A, Durand P, Morand S, et al. Distribution and genetic diversity of Schistosoma haematobium within its bulinid intermediate hosts in Mali. Acta Trop. 1997;66(1):15–26. doi:10.1016/S0001-706X(97)00670-0
46. Afifi M, Jiman-Fatani A, Al-Hussainy N, Al-Rabia M, Bogari A. Genetic diversity among natural populations of Schistosoma haematobium might contribute to inconsistent virulence and diverse clinical outcomes. J Microsc Ultrastruct. 2016;4(4):222. doi:10.1016/j.jmau.2016.04.002
47. Degarege A, Erko B, Mekonnen Z, et al. Comparison of individual and pooled urine samples for estimating the presence and intensity of Schistosoma haematobium infections at the population level. Parasit Vectors. 2015;8(1). doi:10.1186/s13071-015-1205-7.
48. Silva LK, Liu S, Blanton RE. Microsatellite analysis of pooled Schistosoma mansoni DNA: an approach for studies of parasite populations. Parasitology. 2006;132(3):331–338. doi:10.1017/S0031182005009066
49. Shaw SH, Carrasquillo MM, Kashuk C, Puffenberger EG, Chakravarti A. Allele frequency distributions in pooled DNA samples: applications to mapping complex disease genes. Genome Res. 1998;8(2):111–123. doi:10.1101/gr.8.2.111
50. Ezeh C, Yin M, Li H, et al. High genetic variability of Schistosoma haematobium in Mali and Nigeria. Korean J Parasitol. 2015;53(1):129–134. doi:http://dx.doi.10.3347/kjp.2015.53.1.129
51. Glenn TC, Lance SL, McKee AM, et al. Significant variance in genetic diversity among populations of Schistosoma haematobium detected using microsatellite DNA loci from a genome-wide database Internet. Parasit Vectors. 2013;6(1). doi:10.1186/1756-3305-6-300
52. Gower CM, Gouvras AN, Lamberton PH, et al. Population genetic structure of Schistosoma mansoni and Schistosoma haematobium from across six sub-Saharan African countries: implications for epidemiology, evolution and control. Acta Trop. 2013;128(2):261–274. doi:10.1016/j.actatropica.2012.09.014
53. Webster BL, Diaw OT, Seye MM, Webster JP, Rollinson D, Secor WE. Introgressive hybridization of schistosoma haematobium group species in senegal: species barrier break down between ruminant and human schistosomes. Secor WE, editor. PLoS Negl Trop Dis. 2013;7(4):e2110. doi:10.1371/journal.pntd.0002110
54. Moné H, Minguez S, Ibikounlé M, Allienne JF, Massougbodji A, Mouahid G. Natural Interactions between S. haematobium and S. guineensis in the Republic of Benin. Sci World J. 2012;2012:1–8. doi:10.1100/2012/793420
55. Ynt T-B, Webster B, Konan CK, et al. Molecular characterization and distribution of Schistosoma cercariae collected from naturally infected bulinid snails in northern and central Côte d’Ivoire. Parasit Vectors. 2019;12:1–10. doi:10.1186/s13071-019-3381-3
56. MacPherson JM, Eckstein PE, Scoles GJ, Gajadhar AA. Variability of the random amplified polymorphic DNA assay among thermal cyclers, and effects of primer and DNA concentration. Mol Cell Probes. 1993;7(4):293–299. doi:10.1006/mcpr.1993.1043
57. Webster BL, Emery AM, Webster JP, et al. Genetic diversity within Schistosoma haematobium: DNA barcoding reveals two distinct groups. PLoS Negl Trop Dis. 2012;6(10):e1882. doi:10.1371/journal.pntd.0001882
58. Webster BL, Culverwell CL, Khamis IS, Mohammed KA, Rollinson D, Stothard JR. DNA barcoding of Schistosoma haematobium on Zanzibar reveals substantial genetic diversity and two major phylogenetic groups. Acta Trop. 2013;128(2):206–217. doi:10.1016/j.actatropica.2012.06.002
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.