Back to Journals » OncoTargets and Therapy » Volume 12
Genetic association of XRCC5 gene polymorphisms with breast cancer among Jordanian women
Authors AL-Eitan LN , Rababa'h DM, Alghamdi MA , Khasawneh RH
Received 21 June 2019
Accepted for publication 3 September 2019
Published 26 September 2019 Volume 2019:12 Pages 7923—7928
DOI https://doi.org/10.2147/OTT.S220226
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Laith N AL-Eitan,1,2 Doaa M Rababa’h,1 Mansour A Alghamdi,3 Rame H Khasawneh4
1Department of Applied Biological Sciences, Jordan University of Science and Technology, Irbid 22110, Jordan; 2Department of Biotechnology and Genetic Engineering, Jordan University of Science and Technology, Irbid 22110, Jordan; 3College of Medicine, King Khalid University, Abha, Saudi Arabia; 4Department of Hematopathology, King Hussein Medical Center (KHMC), Jordan Royal Medical Services (RMS), Amman 11118, Jordan
Correspondence: Laith N AL-Eitan
Jordan University of Science and Technology, PO Box 3030, Irbid 22110, Jordan
Tel +962 2 720 1000 Ext 23464
Fax +962 2 720 1071
Email [email protected]
Purpose: Breast cancer (BC) is a complex disease that is governed by several different environmental and inherited factors. There are many genes have been linked with BC development by screening specific genetic variants within these genes. In this study, we aim to investigate the correlation between Variable Number Tandem Repeat (VNTR) in XRCC5 gene and BC.
Materials and methods: Polymerase Chain Reaction (PCR) and Gel electrophoresis were used to genotype the XRCC5 gene polymorphism in 200 cases with breast cancer and 200 healthy individuals. All participants were Jordanian women from Arab descents. Clinical and pathological characteristics for BC patients were summarized and categorized according to their medical records.
Results: In this study, we found a strong correlation between the VNTR polymorphism in the XRCC5 gene and BC risk (P-value<0.0001). Remarkably, three different genotypes (2R|2R, 3R|2R and 3R|3R) showed significant association with BC (P-value<0.0001). This study also reported a significant difference in the distribution of allele frequencies between BC patients and healthy individuals (3R; P-value<0.0001 and 2R; P-value<0.001). However, we propose that VNTR of XRCC5 gene did not interfere with BC prognosis.
Conclusion: We speculate that the VNTR of XRCC5 gene may influence BC development. More investigations are needed in this regard to clarify the underlying role of the XRCC5 genetic variant in tumorgenesis including BC development.
Keywords: breast, cancer, XRCC5, genetic polymorphism, prognosis
Background
While breast cancer (BC) can occur in men, it is much more frequent in women, and it is considered the most prevalent and fatal type of cancer affecting women.1–3 In Jordan, BC is the most common female cancer accounting for 38.2% of all cancers BC according to GLOBOCAN reports. Survival rates have increased in recent years due to wider awareness of preventative measures as well as enhanced therapeutic protocols.4 Although the precise cause of BC is still not fully clear, the main risk factors for the disease have been established.5,6 In the past decade, the spotlight of BC research has shifted to the role of inherited risk factors, such as family history and genetic mutations, in the disease’s progression.7–9 It has been reported that the occurrence of certain genetic polymorphisms in critical housekeeping genes contributes to the onset of cancer.10,11
The variable number tandem repeat (VNTR) polymorphism in the X-ray repair cross-complementing protein 5 (XRCC5) gene has been linked to the development of cancer in Iranians and Caucasians.12–14 More specifically, this 160-bp VNTR is located in the XRCC5 promoter region and involves a 21-bp repetitive unit (TGCGCATGCTCGGCGGGAATC).15,16 The XRCC5 gene is located on chromosome 2 and encodes the DNA repair protein Ku80.17 Four different VNTR alleles have been identified: 0R, 1R, 2R and 3R.18
A limited number of studies have investigated the relationship between BC and the XRCC5 VNTR polymorphism. Rajaei et al,18 reported that the 0R/0R genotype led to an increased BC risk compared with the 1R/1R genotype, while Cui et al,14 found that the VNTR in the XRCC5 promoter was associated with an altered BC risk. The primary aim of the current study is to investigate the impact of the XRCC5 promoter VNTR in BC risk as well as its prognosis in the Jordanian population of Arab descent.
Methods
Study population
Ethical approval (Code number: 32/104/2017) from the Intuitional Review Board (IRB) at Jordan University of Science and Technology (JUST) along with written informed consent was obtained for this study and from all its participants, respectively. This study was also conducted in accordance with the Declaration of Helsinki. In total, the 400 samples were utilized for genomic extraction and genotyping: 200 samples from Jordanian BC patients were collected from the Jordanian Royal Medical Services (JRMS) hospital, in addition to 200 samples from randomly selected healthy Jordanian women were recruited from the blood bank at JRMS. Both the patients and the controls were age- and gender-matched and came from the same ethnic background (Arab). In addition, demographic and clinical data of BC samples were extracted from the medical records of DE-identified Arab BC patients.
Genomic extraction and genotyping XRCC5 promoter VNTR polymorphisms
Five milliliters of blood was collected from each participant and DNA was extracted using Wizard® Genomic DNA Purification Kit (Promega Corp., Madison, WI, USA) according to the manufacturer’s instructions. DNA quantity and quality were then assessed using the NanoDropTM (Thermo Scientific, USA). The polymerase chain reaction (PCR) technique coupled with agarose gel electrophoresis was employed to genotype the VNTR in the XRCC5 promoter of each participant. Briefly, 2 µL each of sense (5′AGGCGGCTCAAACACCACAC3′) and antisense (5′CAAGCGGCAGATAGCGGAAAG3′)14 was added to 12.5 µL of ready-to-use master mix, 10 µL of distilled deionized water and 50 ng of DNA. The samples were loaded into a thermal cycler set to the following conditions: initial denaturation at 95° C for 7 mins, 35 cycles of denaturation (95° C for 30 s), annealing (62° C for 30 s), and extension (72° C for 45 s), and a final extension at 72° Cf or 7 mins. The amplicon was separated using 2% agarose gel stained with ethidum bromide (0.07 µg/mL) and placed in TBE buffer. Afterwards, the bands were visualized and documented by a gel image system. The size of the DNA bands was determined through comparison with a 100 bp ladder.
Statistical analysis
Genotypic and allelic frequencies were calculated, and Pearson’s chi-squared test was applied to compare between cases and controls by calculating the P-value.19 The Pearson χ2 test was also used to evaluate the deviation of the Hardy–Weinberg equilibrium (HWE). The HWE equation (p2+2pq+q2+2qr+r2+2ps+2pr+2qs+2sr+s2=1) was used to calculate the estimated multiple allelic (0R, 1R, 2R and 3R) and genotypic (0R\0R, 1R\0R, 1R\1R, 1R\2R, 2R\0R, 2R\2R, 1R\3R, 3R\2R and 3R\3R) frequencies. In this study, Pearson’s Chi-squared and ANOVA tests were also used to perform the genotype-phenotype analyses. A P-value of less than 0.05 was considered statistically significant. The Statistical Package for the Social Sciences (SPSS) version 21.0 was also used to perform all analyses.
Results
VNTR frequency distributions in the XRCC5 promoter among breast cancer (BC) patients and controls
After performing the HWE test for the polymorphism in both cases and controls, it was in accordance with the HWE criteria. Our results involved the following four VNTR alleles: 0R, 1R, 2R and 3R. Each allele possessed a specific molecular weight (Figure 1). Nine different genotypes can result from these alleles. Table 1 summarizes the distribution of these different genotypes and alleles among BC patients and controls.
 |
Table 1 Genotype and allele frequencies of the XRCC5 VNTR in breast cancer patients and healthy individuals |
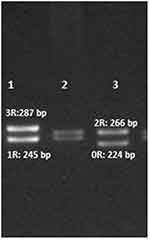 |
Figure 1 Agarose gel electrophoresis of the four alleles of the XRCC5 promoter VNTR polymorphism after PCR amplification. Lanes 1, 2 and 3 show the 3R/1R, 2R/1R and 2R/0R genotypes, respectively. |
As can be seen from Table 1, 35.5% of healthy individuals possessed the 2R\2R genotype, while the 1R\1R genotype was found in 25% of healthy subjects. In contrast, both the 3R\3R and 0R\0R genotypes accounted for only 1% of healthy subjects. According to our findings, the 2R\2R genotype has the highest frequency among controls (71%) while most of the cases had the 1R\2R genotype.
Comparing between case and control genotypes (Table 1), a variation in genotypic frequencies was found between healthy women and BC patients. For instance, 35.5% of healthy subjects had the 2R\2R genotype compared to 19% of BC patients (P-value<0.0001). Moreover, there was a conspicuous disparity in 3R\3R genotype distribution among controls (1%) and patients (8.5%) with P-value<0.0001. Furthermore, significant association of genotypic frequency of 3R\2R genotype among cases and controls with BC risk was observed (P-value<0.0001). The estimated overall P-value for an association between XRCC5 VNTR polymorphism and BC was<0.0001. In terms of the allelic frequency distributions of XRCC5 VNTR polymorphism, a significant difference was observed among cases and controls. Our findings showed a significant association between allelic frequencies of 3R and 2R in the study cohort (P-values<0.0001 and<0.001, respectively).
In addition, we investigated the correlation between certain clinical and pathological parameters of BC patients and the XRCC5 variant. For each parameter, we compared between the groups of patients with different genotypes. Table 2 reviews patients’ clinical characteristics compared with different XRCC5 VNTR genotypes. These parameters include body mass index (BMI), age at first breast cancer diagnosis, age at first pregnancy, age at menarche, age at menopause, co-morbidity, allergies and lifestyle habits such as smoking. In this study, we did not find any correlation between these investigated criteria among BC patients and the VNTR of XRCC5 gene. In addition to clinical features, the existence of any link between pathological features and BC risk was explored. Table 3 summarizes the studied pathological parameters, which included progesterone and estrogen receptors status (PR and ER), human epidermal growth factor receptor 2 (Her2) marker, extent of tumor differentiation (low vs middle and high), tumor size, lymph node involvement and others. In this study, BC was classified into in situ carcinoma and invasive carcinoma. Based on the genetic analysis, we found that 18.9% of patients were diagnosed with in situ carcinoma (2R and/or 3R; 27.8%, 0R and/or 1R; 72.2%) while invasive carcinoma accounted for 81.1% (2R and/or 3R; 35%, 0R and/or 1R; 65.0%).
 |
Table 2 Association of the XRCC5VNTR polymorphism with the clinical characteristics of breast cancer patients |
 |
Table 3 Association of XRCC5 VNTR polymorphism with pathological characteristics of BC patients |
In addition, the genotypes of XRCC5 VNTR were investigated for correlation with different types of BC according to the immunohistochemistry (IHC) profiles of BC patients, these types categorized according to the presence or absence of ER, PR, and HER2 markers (Table 3). However, our results did not show an association between the pathological parameters and XRCC5 VNTR.
Discussion
Breast cancer (BC) comprises a large part of the disease and injury burden for women and poses a direct threat to their health and overall welfare.20,21 From among BC risk factors, the role of genetics in BC development has been widely investigated, and numerous genetic polymorphisms suggested to influence its progression have been screened.22,23 Different genotypes of the VNTR in the XRCC5 gene have been identified by several early studies.24,25 In the present study, ten different genotypes and four alleles of the XRCC5 VNTR polymorphism in its promoter region were assessed for any relation with BC risk in Jordanian females.
XRCC5 gene encodes the Ku80 heterodimer subunit. The Ku80 acts in non-homologous end joining (NHEJ) pathway by binding to DNA double-strand break ends. In addition, this heterodimer subunit plays a key role in homologous recombination (HR). A (VNTR) insertion in XRCC5 promoter region can influence its expression and affect Ku80 synthesis. As a consequence, these changes may affect the NHEJ and HR pathways leading to multiple types of sporadic cancer including BC.14
Previous studies claimed that the VNTR in the XRCC5 gene is vary among the ethnic group.18 Cui et al,14 compared the frequency of all VNTR genotypes among 100 healthy local Caucasian individuals with the genotypes from 535 Caucasian Iranian and 235 Chinese populations. The incidence of 2R\2R genotype in our finding was 35.5% which is far from it in Caucasian (16%), Iranian (2%) and Chinese (5%). Whereas the frequency of homozygous 1R allele among healthy subjects in this study (25%) was compatible with it among Caucasian (22%) and in contrast to it among Iranian and Chinese populations (5% and 2%), respectively. Additionally, the 0R\0R genotype was similar to it among these three population. According to Cui et al,14 the 3R\3R genotype was not detected among Caucasian, Iranian and Chinese. However, only 1% of 3R\3R genotype was detected among healthy individuals in the current Jordanian study. Our results suggest that the genotypic distribution among BC patients and healthy controls was significantly linked to breast cancer (P-value=1.5e–7). A significant difference in the distribution of 2R\2R, 2R\3R and 3R\3R genotypes between cases and controls, proposing that the 2R and 3R may be in association with BC. In the current study, we also observed that the frequencies of the genotypes with low number repeats (0R and 1R) were higher than the genotypes with high number repeats (2R and 3R) among BC cohort which contrasts with the XRCC5 VNTR polymorphism distribution among controls. Depending on the statistically significant results, the distribution of 2R/2R genotype was higher than 3R/3R genotype distribution frequency among BC patients. Furthermore, 2R allele was more frequent than the 3R allele among BC patients. As a consequence, this study suggested that 2R alleles may influence BC increased risk.
Several studies demonstrated the role of XRCC5 VNTR polymorphism in cancer development and progression. Rajaei et al,18 reported that the 3R allele of the XRCC5 VNTR leads to reduced gene expression. Since XRCC5 overexpression has been linked to cancer, an increase in the XRCC5 VNTR repeat number may confer protection against cancer. Subsequent research by Rajaei et al,26 determined that the 0R\0R genotype increased BC risk compared to the 1R\1R genotype, while the 3R\1R genotype decreased BC risk compared to the 1R\1R genotype.
In addition, a group of clinical and pathological criteria thought to be BC risk factors were investigated and statistically tested for any significant association with XRCC5 VNTR. However, we failed to detect any correlation between BC prognosis factors including clinical and pathological features and XRCC5 VNTR polymorphism.
To conclude, the XRCC5 VNTR polymorphism is strongly associated with BC among Jordanian women but not with the clinicopathological parameters. More epidemiological studies are recommended to clarify the underlying role of XRCC5 genetic variant among different ethnic groups.
Ethical approval and informed consent
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Institutional Review Board (IRB) at Jordan University of Science and Technology with ethical code number 32/104/2017. Written informed consent was obtained from all individual participants included in the study.
Data availability
The data sets generated and/or analyzed over the course of the study are not publicly available but are available from the corresponding author on reasonable request.
Acknowledgment
The authors thank the Jordanian Royal Medical Services (JRMS), Amman, Jordan, for approving this study in the first instance and making the clinical data and samples available for the study. This study was funded by the Deanship of Research (RN: 126/2017), Jordan University of Science and Technology (JUST). There was no role of JUST in the design of the study; collection, analysis and interpretation of data; and in writing the manuscript.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Benson JR, Jatoi I. The global breast cancer burden. Future Oncol. 2012;8(6):697–702. doi:10.2217/fon.12.61
2. Harbeck N, Gnant M, Thomssen C. Breast cancer is our global responsibility. Breast Care. 2015;10(6):360. doi:10.1159/000443159
3. Desantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52–62. doi:10.3322/caac.21203
4. Narod SA, Iqbal J, Miller AB. Why have breast cancer mortality rates declined? J Cancer Policy. 2015;5:8–17. doi:10.1016/j.jcpo.2015.03.002
5. Verma R, Bowen RL, Slater SE, Mihaimeed F, Jones JL. Pathological and epidemiological factors associated with advanced stage at diagnosis of breast cancer. Br Med Bull. 2012;103:129–145. doi:10.1093/bmb/lds018
6. Tao Z, Shi A, Lu C, Song T, Zhang Z, Zhao J. Breast cancer: epidemiology and etiology. Cell Biochem Biophys. 2015;72(2):333–338. doi:10.1007/s12013-014-0459-6
7. Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi:10.1038/35020557
8. Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi:10.1073/pnas.191367098
9. Barrow TM, Michels KB. Epigenetic epidemiology of cancer. Biochem Biophys Res Commun. 2014;455:70–83. doi:10.1016/j.bbrc.2014.09.137
10. Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi:10.1038/nature05887
11. Hughes S, Agbaje O, Bowen RL, et al. Matrix metalloproteinase single-nucleotide polymorphisms and haplotypes predict breast cancer progression. Clin Cancer Res. 2007;13:6673–6680. doi:10.1158/1078-0432.CCR-07-0884
12. Alshareeda AT, Negm OH, Albarakati N, et al. Clinicopathological significance of KU70/KU80, a key DNA damage repair protein in breast cancer. Breast Cancer Res Treat. 2013;139:301–310. doi:10.1007/s10549-013-2542-x
13. Rajaei M, Saadat I, Saadat M. Introducing a novel allele for the polymorphism of variable number of tandem repeats in the promoter region of XRCC5. Biochem Biophys Res Commun. 2012;427:503–505. doi:10.1016/j.bbrc.2012.09.085
14. Cui J, Luo J, Kim YC, et al. Differences of variable number tandem repeats in XRCC5 promoter are associated with increased or decreased risk of breast cancer in BRCA gene mutation carriers. Front Oncol. 2016;6:92. doi:10.3389/fonc.2016.00092
15. Jahantigh D, Moghtaderi A, Narooie-Nejad M, et al. Carriage of 2R allele at VNTR polymorphous site of XRCC5 gene increases risk of multiple sclerosis in an Iranian population. Russ J Genet. 2017;53:147. doi:10.1134/S102279541612005X
16. Gorre M, Mohandas PE, Kagita S, et al. Association of XRCC5 VNTR polymorphism with the development of chronic myeloid leukemia. Tumour Biol. 2014;35(2):923–927. doi:10.1007/s13277-013-1120-5
17. Wang G, Wang S, Shen Q, et al. Polymorphisms in XRCC5, XRCC6, XRCC7 genes are involved in DNA double-strand breaks (DSBs) repair associated with the risk of acute myeloid leukemia (AML) in Chinese population, J. Nanjing Med Uni. 2009;23(2):93–99. doi:10.1016/S1007-4376(09)60034-4
18. Rajaei M, Saadat I, Saadat M. The novel allele (3R) of the VNTR polymorphism in the XRCC5 promoter region dramatically decreases the gene expression. Biochem Biophys Res Commun. 2013;430:640–641. doi:10.1016/j.bbrc.2012.10.142
19. Preacher KJ. Calculation for the Chi-square Test: An Interactive Calculation Tool for Chi-square Tests of Goodness of Fit and Independence. New York, NY, USA: Vanderbilt University; 2001.
20. Narod SA, Feunteun J, Lynch HT, et al. Familial breast-ovarian cancer locus on chromosome 17q12-q23. Lancet. 1991;388:82–83.
21. Barnett G, Shah M, Redman K, Easton D, Ponder B, Pharoah P. Risk factors for the incidence of breast cancer: do they affect survival from the disease? J Clin Oncol. 2008;26(20):3310–3316. doi:10.1200/JCO.2006.10.3168
22. Olopade OI, Fackenthal JD, Dunston G, Tainsky MA, Collins F, Whitfield-Broome C. Breast cancer genetics in african americans. Cancer. 2003;97:236–245. doi:10.1002/cncr.11021
23. Turpin E, Bieche I, Bertheau P, et al. Increased incidence of erbb2 overexpression and tp53 mutation in inflammatory breast cancer. Oncogene. 2002;21:7593–7597. doi:10.1038/sj.onc.1205932
24. Wang S, Wang M, Yin S, et al. A novel variable number of tandem repeats (VNTR) polymorphism containing Sp1 binding elements in the promoter of XRCC5 is a risk factor for human bladder cancer. Mutat Res. 2008;638:26–36. doi:10.1016/j.mrfmmm.2007.08.011
25. Jahantigh D, Hosseinzadeh Colagar A. XRCC5 VNTR, XRCC6 −61C>G, and XRCC7 6721G>T gene polymorphisms associated with male infertility risk: evidences from case-control and in silico studies. Int J Endocrinol. 2017. doi:10.1155/2017/4795076
26. Rajaei M, Saadat I, Omidvari S, Saadat M. Association between polymorphisms at promoters of XRCC5 and XRCC6 genes and risk of breast cancer. Med Oncol. 2014;31:885. doi:10.1007/s12032-014-0885-8
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
