Back to Journals » International Journal of General Medicine » Volume 14
Genetic Association of rs10757278 on Chromosome 9p21 and Coronary Artery Disease in a Saudi Population
Authors Bogari N , Dannoun A, Athar M, Elkhateeb O, Porqueddu M, Allam R , Alamanni F
Received 19 January 2021
Accepted for publication 6 April 2021
Published 5 May 2021 Volume 2021:14 Pages 1699—1707
DOI https://doi.org/10.2147/IJGM.S300463
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Scott Fraser
Neda Bogari,1 Anas Dannoun,1 Mohammad Athar,1,2 Osama Elkhateeb,3,4 Massimo Porqueddu,5,6 Reem Allam,7 Francesco Alamanni6
1Department of Medical Genetics, Faculty of Medicine, Umm Al-Qura University, Makkah, Kingdom of Saudi Arabia; 2Science and Technology Unit, Umm Al-Qura University, Makkah, Kingdom of Saudi Arabia; 3Department of Cardiology, King Abdullah Medical City Makkah, Makkah, Kingdom of Saudi Arabia; 4Department of Cardiology, Dalhousie University Halifax, Nova Scotia, Canada; 5Department of Cardiac Surgery, King Fahd Armed Forces Hospital, Jeddah, Kingdom of Saudi Arabia; 6Department of Cardiac Surgery, Monzino Heart Center, University of Milan, Milano, Italy; 7Clinical Pathology Department, Faculty of Medicine, Zagazig University, Sharkia, Egypt
Correspondence: Neda Bogari
Department of Medical Genetics, Faculty of Medicine, Umm Al-Qura University, Makkah, Kingdom of Saudi Arabia
Tel + 966 25270000 Ext:5057-4168
Fax +966 25272155
Email [email protected]
Reem Allam
Clinical Pathology Department, Faculty of Medicine, Zagazig University, Sharkia, Egypt
Email [email protected]
Purpose: Coronary artery disease (CAD) is one of the most important leading causes of morbidity and mortality worldwide. Few studies have been carried out in the Saudi population regarding the association of rs10757278 polymorphism with CAD. This study aimed to investigate the association of the rs10757278 polymorphism with CAD in Saudi population.
Materials and Methods: In this case-control study, we recruited 437 patients with CAD and 251 cross-matched healthy controls and performed polymorphism genotyping for rs10757278 using a polymerase chain reaction followed by a restriction fragment length polymorphism analysis.
Results: The G allele (OR-1.44; 95% CI: 1.15– 1.80; p=0.001), as GG (OR-2.13; 95% CI: 1.35– 3.36; p=0.0009), in the dominant (OR-1.47; 95% CI: 1.03– 2.10; p=0.03) and recessive mode (OR-1.84; 95% CI: 1.26– 2.70; p=0.001) of inheritance showed a high-risk association. A disease stratified risk analysis was conducted and comparisons were made using an ANOVA analysis. Diabetes showed a risk association (p=0.001). However, a regression analysis confirmed that for the CAD cases, there was an association between the GG genotype and diabetes (p=0.005).
Conclusion: The results of this study suggest that the polymorphism rs10757278 is related to a high risk of CAD in a Saudi population.
Keywords: coronary artery disease, 9p21, rs10757278, gene polymorphism and Saudi population
Introduction
Coronary artery disease (CAD) is a chronic, complex, multifactorial, continuous inflammatory condition that results from the accumulation of atheromatous plaques in intima layer of the coronary arteries. CAD is responsible for numerous acute coronary syndromes and is the main cause of death in the Western hemisphere.1 CAD is a multifactorial illness that is affected by both acquired and inherited factors. Certain risk factors for CAD are modifiable, such as lifestyle changes and medication management.3 However, there are numerous genetic and environmental risk factors contributing to CAD with high mortality and morbidity rate in most developed and developing countries.4 Myocardial infarction and angina are considered coronary vascular diseases (CVD), reported in 31% of the population worldwide.5 Specifically, the World Health Organization (WHO) reported that around 17.9 million people succumbed to CVD in 2016.6 Thus, a better understanding of the various CVDs is of great interest.
CAD is associated with conventional risk factors, including smoking, hypertension (HTN), diabetes mellitus (DM), poor diet and advanced age.7 However, the etiology of CAD remains obscure so far.8 The lesion incidence and long-term prognosis of CAD is variable. For example, patients with similar lesions may become stable or progressive and receive similar treatments. In addition, a small proportion of CAD patients show none of the classical risk factors. Thus, these observations support the unclear etiology. However, researchers suggest that genetic factors also contribute not only to the development of atherosclerosis but also to the development of the classical risk factors.9 In fact, CAD family aggregation has been recognized for quite some time, and the genetic predisposition to CAD is confirmed by conclusive evidence. For example, twin studies reveal that the estimates of heritability for CAD range from 41 to 77%, and the significance of genetic factors in CAD led to the extensive discovery of significant candidate genes and multiple small nucleotide polymorphisms (SNPs) being linked to this condition.10,11 The exact mechanism that underlies the impact of polymorphisms on the pathogenesis of CAD is not fully understood. However, polymorphisms in genes related to inflammation, lipid and glucose metabolism, blood, coagulation and homocysteine can affect the susceptibility to CAD.13 Thus, genotyping specific SNPs within a potential CAD-related gene is an essential and effective method to detect genetic risk markers, and numerous significant CAD-related SNPs have been reported.14 In particular, one SNP at loci 9p21.3 has been identified as a global population hotspot and is linked to CAD.15 Helgadottir et al16 identified the SNP variant of rs10757278 on chromosome 9p21.3 using genome-wide association studies (GWAS) that included both myocardial infarction and CAD patients. In addition, a meta-analysis by Hu et al17 reported that the rs10757278 polymorphism was significantly linked to CAD. Nevertheless, the rs10757278 polymorphism studies related to CAD are very few, if present, in Saudi population and this area of research is not fully elucidated. Moreover, the vast area of Saudi Arabia and far distances among its governorates necessitates wider research on CAD in different localities inside Saudi Arabia. Therefore, the current study aimed to investigate the genetic association of the rs10757278 polymorphism in Saudi patients with confirmed CAD.
Materials and Methods
Recruitment of the Participants
This case-control study included 688 patients, consisting of 437 CAD subjects and 251 healthy subjects from King Abdullah Medical City (KAMC) and Al-Noor specialized hospitals in the Jeddah and Mecca regions of Saudi Arabia. Sample size was calculated by statistical expert using open Epi version 6 program, at 80% power of study, 95% confidence level,18 as the total number of patients with ischemic heart diseases mounts to 167,499 people according to the Ministry of Health (MOH) Statistical Yearbook and the mutant genotype frequency in Eastern area of Saudi Arabia was found to be around 50%.19 We add 10% sample to compensate for drop-out and missed cases. The research protocol was approved by the Umm Al-Qura (43430838–05/05/1434H) and KAMC (13/043) institutes. Before enrolling in the study, all the participants signed an informed consent form. The study was conducted as per the Helsinki Declaration. Our recent publication20 described the selection of the cases and controls in detail. The inclusion criteria for the CAD cases were selected based on full assessments in the hospital premises. In the age range of 30–85 years of age, the reported CAD cases were 50% higher than coronary artery stenosis. The CAD exclusion criteria were non-Saudi nativity, malignant diseases and/or concomitant inflammatory diseases. The type 2 diabetes mellitus (T2DM) cases were defined based on the American Diabetes Association criteria.22–24 The 251 apparently healthy controls were chosen among blood donors visiting same hospitals with no family history of CAD, heart pain, stroke or peripheral vascular diseases. Cross-matching was done according to age, sex and residence. The control subjects’ exclusion criteria were malignancy and/or with autoimmune diseases.
Blood and Serum Analysis
Total cholesterol (TC), triglycerides (TG), high-density lipoprotein-cholesterol (HDL - c) and low-density lipoprotein-cholesterol (LDL - c) were obtained from the enrolled (n=688) subjects, and 5 mL of coagulant blood was transferred for the biochemical analysis of the fasting blood glucose (FBG) and the lipid profile analysis (LDL - c).25 The samples for FBG were collected after an overnight fasting.
Molecular Analysis
A total of 5 mL of EDTA anticoagulant blood was used to isolate the genomic DNA using the Promega Wizard Genomic DNA purification kit as recommended by the manufacturers, and it was stored at −20°C until further use. In order to measure the quality and quantity of the genomic DNA, it was analyzed using a NanoDrop Spectrophotometer. The genotyping for rs10757278 was performed using an amplification refractory mutation system-polymerase chain reaction (ARMS-PCR) with the following primer sequences:
Forward Outside: 5ʹGCTGTCCAGCAGCAGCAGCAGCTCTCT-3’;
Reverse Outside: 5ʹAAGGGCATTAAGGGTGGTGGTAGACAA-3’;
A allele Primer: 5ʹACTACTCTGTCTGCTGCTGCTT-3’; and
G Allele Primer: 5ʹAGTCAGGTGTGTGCATTCGGGAA-3’.
The PCR amplification was carried out on a Veriti Life Technology Thermal Cycler. The GoTaq Green Master Mix (Promega Co. Ltd) was used for the reaction, and the conditions were an initial denaturation at 95°C for 5 min, a subsequent denaturation at 95°C for 30 s, annealing at 60°C for 1 min, extension at 72°C for 45 s and a final extension at 72°C for 7 min for 35 cycles and a final 4°C hold. The undigested PCR products were run on a 3% ethidium bromide agarose gel. A PCR fragment of 445 bp was observed along with an A allele fragment of 263 bp and a G allele fragment of 238 bp. The AA genotype produced bands of 445/263 bp, AG showed bands at 445/263/238 bp and GG had bands of 445/238 bp (Figure 1). Ten percent of the study samples were validated by DNA/sanger sequencing for quality control. The findings from the study of ARMS-PCR and sequencing were 100% concordant, and the representative chromatograms of the wild-type (AA), heterozygous (AG) and homozygous (GG) genotypes (Figure 2).26,27
 |
Figure 1 The rs10757278 genotyping for the G/A alleles using ARMS-PCR. Lane 1: 50 bp ladder. Lane 2–6: GG genotypes. Lane 7–11: AA genotypes. Lane 12–20: AG genotypes. |
 |
Figure 2 Representation of the validation by DNA sequencing for rs10757278. |
Statistical Analysis
The statistical data analyses were performed using version 21 of SPSS (Chicago, IL, United States). The continuous variables were tested for normality using a Kolmogorov–Smirnov test. The normally distributed variables are expressed as the mean ± standard deviation (SD) and were assessed using a Student’s t-test. The non-normally distributed variables are expressed as the median ± standard error of mean (SEM) and were assessed using Mann–Whitney U-tests. The nominal variables are presented as the number and percentage frequency and were assessed using a Pearson’s Chi-square (χ2) test. For the CAD instances, the genotypes were evaluated for Hardy Weinberg-equilibrium (HWE), and a chi-square (χ2) analysis was used for the controls. The genotype and allele frequencies were calculated using Openepi software, and the cases and controls were evaluated using chi-square, odds ratio (OR) and 95% confidence intervals (CI’s). In an independent group, the data for 3 genotypes were analyzed using the means of a one-way variance analysis (ANOVA) or Kruskal–Wallis with a post-hoc test for significance. In addition, a multiple regression analysis was performed using the GG genotypes and the associated risk factors. A P-value <0.05 was considered statistically significant.28,29
Results
Clinical and Biochemical Data
A case-control study was conducted that consisted of 437 CAD cases and 251 controls in a Saudi population. The patients with CAD had a mean age (SD) of 59.04 (5.19) and 66% were males, 57.2% were diabetic and 68% hypertensive. The subjects in the control group were age-matched and 45.4% were males. The lifestyle and clinical characteristics of the participants involved in the study are shown in (Table 1). There was a highly significant difference between the cases and controls with regard to weight, BMI, smoking, diabetes, hypertension, stroke, heart failure, blood glucose level and all of the lipid parameters evaluated.
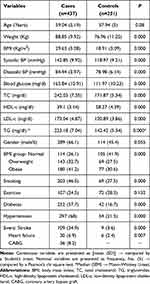 |
Table 1 Characteristics of the Study Population Cases and Controls |
Genotype and Allele Frequencies
For the CAD cases and controls, the genotype distribution showed no evidence of divergence from HWE (p>0.05). The distribution of both alleles and the genotype frequencies of the polymorphism rs10757278 at the 9p21.3 locus for the CAD cases and controls is shown in Table 2 and Figure 3. Among the CAD cases, the frequencies of the AA, AG and GG genotypes were 21.7%, 49% and 29.3% and were 29.1%, 52.6% and 18.3% for the control subjects, respectively. The mutant GG genotype significantly doubled the risk of CAD among the cases when compared to the controls (OR-2.13; 95% CI (1.35–3.36); p=0.0009). Also, the subjects that carried the G allele was more 1.44 times likely to develop CAD (OR-1.44; 95% CI (1.15–1.80); p=0.001). The AA genotype was protective against CAD when compared to the GG and AG genotypes (OR-0.6773; 95% CI (0.475–0.965); p=0.03). While for the recessive model, the GG genotype significantly increases the risk of CAD by 1.84 times compared to the AA and AG genotypes (OR-1.84; 95% CI (1.26–2.70); p=0.001). There was no association between CAD and the heterozygous genotype when it was compared to either the wild-type genotype alone or inside the co-dominant genetic model (p=0.250 and 0.360, respectively).
 |
Table 2 Comparison of the Different Mode of Inheritance of the Genotypes and Allele Frequencies with the rs10757278 Polymorphism in CAD Cases and Control Subjects |
 |
Figure 3 Genotype and allele frequencies of the CAD cases and control subjects. |
Genotype Distribution Among the Various Risk Factors
To study the distribution of the genotype among various risk factors, an ANOVA and Kruskal–Wallis analyses were performed, and the results are displayed in Table 3. In comparison, both physical activity and diabetes, which are modifiable risk factors for CAD, were statistically significant among the three genotypes (p<0.05). In addition, the post-hoc test revealed that the difference was attributed to the mutant GG genotype. Gender is one of the non-modifiable risk factors which, along with the other modifiable risk factors, such as HTN and DM, was not significant among the various genotypes (p> 0.05).
 |
Table 3 Risk Factor Distribution Across Genotypes |
Characteristics of the Lipid Risk Factors Among the Genotypes
The mean differences among different genotypes were not statistically significant regarding lipid parameters, triglycerides, total cholesterol, LDL-c and HDL-c (Table 3).
Independent Associations with CAD
To minimize the effect of the confounding risk factors, a multiple regression analysis was conducted with the GG genotype and the variable risk factors, including DM, HTN, physical activity and the lipid parameters, as listed in Table 4. The rs10757278 polymorphism was an independent risk factor for CAD. The multiple regression analysis, along with the associated risk factors, indicated that GG genotype, DM, HTN, TC, LDL-c and TG showed significant associations (p<0.05).
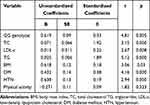 |
Table 4 Multiple Regression Analysis of the Associated Risk Factors |
Discussion
In this case-control study, the rs10757278 polymorphism on the 9p21 locus was confirmed to be associated with CAD in Saudi patients. The GG genotype, G allele and the dominant and recessive models of rs10757278 polymorphism were strongly associated with CAD in Saudi population. The main findings of the present study were as follows: (1) this was the first study conducted in a Saudi population with CAD assessing the rs10757278 polymorphism with separate inheritance and allele modes; (2) several population studies of European and Asian origin consider the GG genotype of the rs10757278 polymorphism to be a risk of CAD, which was confirmed in Saudi Arabia; (3) the trigger allele G for the SNP rs10757278 increased the risk of CAD with an OR of 1.44; (4) in Saudi CAD subjects, the rs10757278 polymorphism was strongly associated with the GG genotype, the G allele and a dominant and recessive mode of inheritance and (5) a regression analysis revealed that, together the GG genotype, DM, HTN and lipid parameters, as its associated risk factors, were significantly correlated in CAD patients.
CAD is a complex, multifactorial cardiovascular disorder that is caused by an interaction between genetic and environmental risk factors. GWAS provides an excellent way for a complete genome assessment of the genomic variants associated with common, complex disorders, such as CAD.30 Globally, CAD is reported as one of the most frequent causes of death, affecting 3.8 million men and 3.4 million women.31 GWAS demonstrates a significant relationship in different populations between the 9p21 nucleotide sequence and the incidence of CAD. This locus consists of ANRIL, a non-coding sequence of antisense RNA and no protein-coding genes.17 Advances in molecular and biochemical approaches have revised our knowledge of CVD and CAD-causing metabolic disorders. In addition, GWAS and meta-analysis studies also identify several disease-causing genes that are further correlated and linked to the molecular basis of CAD diseases.32 At the 9p21 locus, which lies outside of annotated genes, the genetic and molecular basis of CAD risk heterogeneity in the genetic locus is unclear. A study conducted by Broadbent et al34 reported that a high-risk haplotype of 9p21 collocates within a large non-coding antisense RNA gene, which is expressed in tissues affected by atherosclerosis as well as cell types that act as an important regulatory element for growth.
The rs10757278 polymorphism shows both positive35,36 and negative associations.31 However, Hu et al17 and Xu et al37 conducted meta-analysis studies and concluded that the rs10757278 polymorphism might serve as a genetic marker for CAD. Our research results are in line with the Niemiec et al35 study, which showed that rs10757278, independent of traditional risk factors, affected CAD susceptibility.16,38–40 In 2011, the rs10757278 polymorphism was first evaluated in a PAGE analysis to assess its association in two populations according to linkage disequilibrium, and the correlation coefficient high. In addition, a large-scale meta-analysis study suggests that the rs10757278 polymorphism is again significantly associated with MI.42 In one of the Framingham Heart studies, a 13-Kb locus on chromosome 9 (9p21) was associated with MI, stroke and fatal heart disease outcome. This locus consists of several SNPs, including rs1333049 and rs10757278.43 Importantly, the rs10727578 polymorphism has been connected to atherosclerosis, and this polymorphism is associated with progression in the severity and degree of atherosclerosis.40 Scheffold et al indicated that rs10757278 had a greater impact on the risk of CAD in patients with a positive family history.
The strength of this study is that it included 437 Saudi patients diagnosed with CAD, and we included controls that were matched for age and sex. Another strength of this study is that we conducted confirmation analyses to support the correct genotyping results. One limitation of the current study was that it was carried out only with a single SNP using an ARMS-PCR analysis. The final limitation of this study is that this study was carried out in Saudi subjects confirmed with CAD in the western region of Saudi Arabia and to better interpret the results we will need to properly authenticate Saudi expatriate CAD patients.
In conclusion, the present study indicated that allele, genotypes and different genetic inheritance modes were associated with CAD cases compared to controls. Through the regression analysis, the GG genotype was strongly associated with CAD. The current study confirmed the association of the rs10757278 polymorphism with patients diagnosed with CAD in a Saudi population. Future studies conducting analysis of CAD severity association with rs10757278 polymorphism and other genetic polymorphisms in CAD patients with different clinical presentations are being worked on.
Acknowledgment
The authors would like to thank the Deanship of Scientific Research at Umm Al-Qura University for supporting this work by Grant Code 20UQU0068DSR. We are thankful to Dr. Ahmed Shawky and the staff of the Science and Technol- ogy Unit (STU) and Deanship of Scientific Research at Umm-Al-Qura University, Makkah, and Innovation (MAARIFAH)-King Abdulaziz City for Science and Technology-the Kingdom of Saudi Arabia for their support, as well as the Monzino Heart Center, the University of Milan, Milan, Italy, the King Fahd Armed Forces Hospital and King Abdullah Medical City Makkah.for their continuous support. The authors also acknowledge the support of Mr. Abdulmoniem Gowda, Dr. Ahmed Fawzy, Dr. Dareen Ibrahim Rednah, Dr. Saad Alghamdi, Dr. Khalid Faruqui, Dr. Enas Alharthi and Dr. Elaf Almatrouk. We all are deeply appreciative of Mr. Soud Abdulraof A Khogeer, Mr. Abdulmonim Gowda, Mr. Sami Kalantan, Mr. Mustafa N Bogari, Miss Dema N Bogari and Mr. Ehab Melibary.Mr Abdulaziz Alhussini and Mr. Soud Abdulraof A. Khogeer from the Department of Biochemistry, Umm Al-Qura University, Makkah, KSA.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Rocha LO, Rocha E, Succi GDM, Brito junior RBD. Association between periodontitis, genetic polymorphisms and presence of coronary artery disease in Southern Brazil. Arq Bras Cardiol. 2020;114(2):268–272. doi:10.36660/abc.20180296
2. Khattak MF, Horne S. The use of CT coronary angiography and CT fractional flow reserve in the investigation of patients with suspected coronary artery disease. Cureus. 2020;12(5).
3. Pourkeramati A, Mehrjardi EZ, Tezerjani MD, Seifati SM. Association of GSTP1, GSTT1 and GSTM1 gene variants with coronary artery disease in Iranian population: a case–Control Study. Int J Gen Med. 2020;13:249–259. doi:10.2147/IJGM.S252552
4. Luo Z, Zhang T, Wang S, He Y, Ye Q, Cao W. The Trp64Arg polymorphism in β3 adrenergic receptor (ADRB3) gene is associated with adipokines and plasma lipids: a systematic review, meta-analysis, and meta-regression. Lipids Health Dis. 2020;19(1):1–12. doi:10.1186/s12944-020-01290-y
5. Jha CK, Mir R, Elfaki I, et al. Evaluation of the association of omentin 1 rs2274907 a> t and rs2274908 g> a gene polymorphisms with coronary artery disease in Indian population: a case control study. J Pers Med. 2019;9(2):30. doi:10.3390/jpm9020030
6. Tu G, Fang Z, Zhao Y, Wu Q. Association of +138I/D and Lys198Asn polymorphisms in the endothelin-1 gene with early onset of coronary artery disease among the Chinese Han population. Med Sci Monit. 2020;26:26. doi:10.12659/MSM.921542
7. Hou J, Deng Q, Guo X, Deng X, Zhong W, Zhong Z. Association between apolipoprotein E gene polymorphism and the risk of coronary artery disease in Hakka postmenopausal women in southern China. Lipids Health Dis. 2020;19(1):1–7. doi:10.1186/s12944-020-01323-6
8. Lu S, Wang Y, Wang Y, et al. The IL-6 rs1800795 and rs1800796 polymorphisms are associated with coronary artery disease risk. J Cell Mol Med. 2020.
9. Liu R, Song L, Jiang L, et al. Susceptible gene polymorphism in patients with three-vessel coronary artery disease. BMC Cardiovasc Disord. 2020;20(1):1–8. doi:10.1186/s12872-019-01312-3
10. Wang Y, Wu B, Zhang M, Miao H, Sun J. Significant association between rs28362491 polymorphism in NF-κB1 gene and coronary artery disease: a meta-analysis. BMC Cardiovasc Disord. 2020;20(1):1–8. doi:10.1186/s12872-020-01568-0
11. Khan IA, Poornima S, Jahan P, Rao P, Hasan Q. Type 2 diabetes mellitus and the association of candidate genes in Asian Indian population from Hyderabad, India. J Clin Diagn Res. 2015;9(11):GC01. doi:10.7860/JCDR/2015/14471.6855
12. Matam K, Khan IA, Hasan Q, Rao P. Coronary artery disease and the frequencies of MTHFR and PON1 gene polymorphism studies in a varied population of Hyderabad, Telangana region in South India. J King Saud Univ Sci. 2015;27(2):143–150. doi:10.1016/j.jksus.2014.09.002
13. Hassanzadeh-Makoui R, Razi B, Aslani S, Imani D, Tabaee SS. The association between Matrix Metallo-proteinases-9 (MMP-9) gene family polymorphisms and risk of Coronary Artery Disease (CAD): a systematic review and meta-analysis. BMC Cardiovasc Disord. 2020;20(1):1–15. doi:10.1186/s12872-020-01510-4
14. Rehman K, Jabeen K, Awan FR, Hussain M, Saddique MA, Akash MSH. Biochemical investigation of rs1801282 variations in PPAR‐γ gene and its correlation with risk factors of diabetes mellitus in coronary artery disease. Clin Exp Pharmacol Physiol. 2020;47(9):1517–1529. doi:10.1111/1440-1681.13339
15. Omidi S, Ebrahimzadeh F, Kalayinia S. 9P21. 3 locus; An important region in coronary artery disease: a panel approach to investigation of the coronary artery disease etiology. Int J Cardiovasc Pract. 2019;4(2):21–35. doi:10.29252/ijcp-25001
16. Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316(5830):1491–1493. doi:10.1126/science.1142842
17. Hu L, Su G, Wang X. The roles of ANRIL polymorphisms in coronary artery disease: a meta-analysis. Biosci Rep. 2019;39(12). doi:10.1042/BSR20181559
18. Dean AG, Dean JA. Epi Info, Version 6: A Word-Processing, Database, and Statistics Program for Public Health on IBM-Compatible Microcomputers. GA: Epidemiology Program Office, Centers for Disease Control and Prevention; 1994.
19. AbdulAzeez S, Al-Nafie AN, Al-Shehri A, et al. Intronic polymorphisms in the CDKN2B-AS1 gene are strongly associated with the risk of myocardial infarction and coronary artery disease in the Saudi population. Int J Mol Sci. 2016;17(3):395. doi:10.3390/ijms17030395
20. Bogari NM, Aljohani A, Dannoun A, et al. Correlation between rs320 variant in the lipoprotein lipase gene and presence of coronary artery disease and stroke among the Saudi population. Saudi J Biol Sci. 2020;27(8):2018–2024. doi:10.1016/j.sjbs.2020.06.029
21. Alharbi KK, Hussain T, Alharbi FK, et al. Apolipoprotein C3 gene variants and risk of developing type 2 diabetes in Saudi subjects. Metab Syndr Relat Disord. 2015;13(7):298–303. doi:10.1089/met.2015.0022
22. Khan IA, Vattam KK, Jahan P, Mukkavali KK, Hasan Q, Rao P. Correlation between KCNQ1 and KCNJ11 gene polymorphisms and type 2 and post-transplant diabetes mellitus in the Asian Indian population. Genes Dis. 2015;2(3):276–282. doi:10.1016/j.gendis.2015.02.009
23. Alharbi KK, Khan IA, Al-Daghri NM, et al. ABCA1 C69T gene polymorphism and risk of type 2 diabetes mellitus in a Saudi population. J Biosci. 2013;38(5):893–897. doi:10.1007/s12038-013-9384-x
24. Khan IA, Jahan P, Hasan Q, Rao P. Genetic confirmation of T2DM meta-analysis variants studied in gestational diabetes mellitus in an Indian population. Diabetes Metab Syndr. 2019;13(1):688–694. doi:10.1016/j.dsx.2018.11.035
25. Alharbi KK, Khan IA, Munshi A, Alharbi FK, Al-Sheikh Y, Alnbaheen MS. Association of the genetic variants of insulin receptor substrate 1 (IRS-1) with type 2 diabetes mellitus in a Saudi population. Endocrine. 2014;47(2):472–477. doi:10.1007/s12020-014-0177-2
26. Alharbi KK, Al-Sulaiman AM, Bin Shedaid MK, et al. MTNR1B genetic polymorphisms as risk factors for gestational diabetes mellitus: a case-control study in a single tertiary care center. Ann Saudi Med. 2019;39(5):309–318. doi:10.5144/0256-4947.2019.309
27. Khan IA, Jahan P, Hasan Q, Rao P. Relationship between PTEN and gestational diabetes in Asian Indians womens. J Health Spec. 2015;3(3):184. doi:10.4103/1658-600X.159910
28. Khan IA, Movva S, Shaik NA, et al. Investigation of Calpain 10 (rs2975760) gene polymorphism in Asian Indians with gestational diabetes mellitus. Meta Gene. 2014;2:299–306. doi:10.1016/j.mgene.2014.03.001
29. Shahsavari G, Nouryazdan N, Adibhesami G, Birjandi M. Genetic associations and serum paraoxonase levels with atherosclerosis in western Iranian patients. Mol Biol Rep. 2020;47(1):1–8. doi:10.1007/s11033-019-04608-x
30. Yaghoubi Hariri F, Salahshourifar I, Zare KS. Association between coronary artery disease and rs10757278 and rs1333049 polymorphisms in 9p21 locus in Iran. Rep Biochem Mol Biol. 2020;9(1):58–63. doi:10.29252/rbmb.9.1.58
31. Sirotina S, Ponomarenko I, Kharchenko A, et al. A novel polymorphism in the promoter of the CYP4A11 gene is associated with susceptibility to coronary artery disease. Dis Markers. 2018;2018:2018. doi:10.1155/2018/5812802
32. Broadbent HM, Peden JF, Lorkowski S, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17(6):806–814. doi:10.1093/hmg/ddm352
33. Niemiec P, Gorczynska-Kosiorz S, Iwanicki T, et al. The intronic polymorphisms polymorphism of the 9p21. 3 locus is associated with premature coronary artery disease in Polish patients. Genet Test Mol Biomarkers. 2012;16(9):1080–1085. doi:10.1089/gtmb.2012.0046
34. Foroughmand AM, Nikkhah E, Galehdari H, Jadbabaee MH. Association study between coronary artery disease and rs1333049 and rs10757274 polymorphisms at 9p21 locus in South-West Iran. Cell J. 2015;17(1):89.
35. Xu L-B, Zhang Y-Q, Zhang -N-N, et al. Rs10757274 gene polymorphisms in coronary artery disease: a systematic review and a meta-analysis. Medicine. 2020;99(3):e18841. doi:10.1097/MD.0000000000018841
36. Koch W, Hoppmann P, Schömig A, Kastrati A. Variations of specific non-candidate genes and risk of myocardial infarction: a replication study. Int J Cardiol. 2011;147(1):38–41. doi:10.1016/j.ijcard.2009.07.028
37. Lemmens R, Abboud S, Robberecht W, et al. Variant on 9p21 strongly associates with coronary heart disease, but lacks association with common stroke. Eur J Hum Genet. 2009;17(10):1287–1293. doi:10.1038/ejhg.2009.42
38. Patel RS, Su S, Neeland IJ, et al. The chromosome 9p21 risk locus is associated with angiographic severity and progression of coronary artery disease. Eur Heart J. 2010;31(24):3017–3023. doi:10.1093/eurheartj/ehq272
39. Chen G, Fu X, Wang G, Liu G, Bai X. Genetic variant rs10757278 on chromosome 9p21 contributes to myocardial infarction susceptibility. Int J Mol Sci. 2015;16(5):11678–11688.
40. Larson MG, Atwood LD, Benjamin EJ, et al. Framingham Heart Study 100K project: genome-wide associations for cardiovascular disease outcomes. BMC Med Genet. 2007;8(S1):S5. doi:10.1186/1471-2350-8-S1-S5
41. Xie F, Chu X, Wu H, et al. Replication of putative susceptibility loci from genome-wide association studies associated with coronary atherosclerosis in Chinese Han population. PLoS One. 2011;6(6):e20833. doi:10.1371/journal.pone.0020833
42. Scheffold T, Kullmann S, Huge A, et al. Six sequence variants on chromosome 9p21. 3 are associated with a positive family history of myocardial infarction: a multicenter registry. BMC Cardiovasc Disord. 2011;11(1):9. doi:10.1186/1471-2261-11-9
43. Çakmak HA, Bayoğlu B, Durmaz E, et al. Evaluation of association between common genetic variants on chromosome 9p21 and coronary artery disease in Turkish population. Anatol J Cardiol. 2015;15(3):196. doi:10.5152/akd.2014.5285
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
