Back to Journals » The Application of Clinical Genetics » Volume 12
Genetic Analysis Of ABCA1 Gene Of Primary Glaucoma In Jordanian Arab Population
Authors Alkhatib R , Abudhaim N, AL-Eitan L , Abdo N, Alqudah A, Aman H
Received 29 April 2019
Accepted for publication 11 September 2019
Published 4 October 2019 Volume 2019:12 Pages 181—189
DOI https://doi.org/10.2147/TACG.S213818
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Martin Maurer
Rami Alkhatib,1 Nada Abudhaim,1 Laith AL-Eitan,2 Nour Abdo,3 Asem Alqudah,4 Hatem Aman1
1Department of Biotechnology and Genetic Engineering, Jordan University of Science and Technology, Irbid 22110, Jordan; 2Department of Applied Biological Sciences, Jordan University of Science and Technology, Irbid 22110, Jordan; 3Department of Public Health, Faculty of Medicine, Jordan University of Science and Technology, Irbid 22110, Jordan; 4Department of Ophthalmology, Faculty of Medicine, Jordan University of Science and Technology, Irbid 22110, Jordan
Correspondence: Rami Alkhatib
Department of Biotechnology and Genetic Engineering, Jordan University of Science and Technology, Irbid 22110, Jordan
Tel +9622776319493
Email [email protected]
Background: Glaucoma is a neurodegenerative disease that leads to progressive loss of retinal ganglion cells, causing irreversible visual field defects. At the present time, glaucoma is clinically defined but the exact etiology is unknown. The aim of this study is to genotype rs2472493 and rs2487032 SNIPs within ABCA1 gene in 52 Jordanian Arab patients with primary glaucoma and 96 control subjects, and also to investigate the genetic association of these SNPs with primary glaucoma.
Methods: DNA was extracted from both patients and controls according to a well-established procedure. Then, DNA was amplified by PCR using specific primers for this gene. Analysis of polymorphisms was carried out by using DNA sequencing genotyping method.
Results: The results showed that the two SNPs (rs2472493 and rs2487032) located upstream of ABCA1 gene have no significant associations with primary glaucoma disorder (P > 0.05).
Conclusion: This study is the first of its kind to reveal no genetic association between ABCA1 gene and primary glaucoma disorder in Jordanian population of Arab descent.
Keywords: ABCA1, glaucoma, polymorphisms, Jordanian Arab, primers
Introduction
Glaucoma is a neurodegenerative disease that leads to progressive loss of retinal ganglion cells, causing irreversible visual field defects.1–3 It is the second leading cause of irreversible blindness in the world after cataract.4 Until now, the exact etiology of glaucoma is unknown, but it is clinically defined by the presence of characteristic changes in the optic disc and defects in the visual field correspondingly.5 It is affecting more than 60 million people over the world, disproportionately affecting mainly women and Asians.4,6 Glaucoma is a genetically heterogeneous disorder which results from the interaction between several genetic and environmental factors. Therefore, both genetic and environmental influences determine the risk of this disorder.7 Glaucoma has been classified into two major types according to its cause.8 The first type is the primary glaucoma which involved several subtypes. The most prevalent form is primary open angle glaucoma (POAG) which occurs at the age of 35 years and above. Secondly, primary congenital glaucoma (PCG) is considered the most prevalent childhood glaucoma that occurs in early infancy or at birth in the age up to 3 years. Moreover, it is a major cause of blindness in this young population and it is inherited commonly as an autosomal recessive trait. Thirdly, juvenile-onset open angle glaucoma (JOAG) occurs in early onset in the age of 10–35 years and it is inherited as autosomal dominant trait. Finally, primary angle closure glaucoma (PACG) is considered as the second prevalent form of glaucoma after POAG.9
The second type of glaucoma is resulting from injury, inflammation, tumor, or in advanced cases of cataract or diabetes which called as secondary glaucoma.10 The main pathology in glaucoma is caused by elevated intraocular pressure (IOP). Several genetics studies including candidate genes, linkage analysis studies, and genome wide association studies (GWAS) have identified several loci associated with the development of POAG.7,11 A candidate gene study including CYP1B1 gene suggested that the CYP1B1 was involved in a proportion of POAG and PACG cases, and the similar haplotype background of mutations is indicative of their common origin across different glaucoma phenotypes.12 Classical linkage approaches in POAG families allowed the delineation of 14 loci (GLC1A-1N).13 Thus, glaucoma-associated genes have been identified in three of them including myocilin (MYOC/GLC1A),7 optineurin (OPTN/GLC1E),14 and WD repeat domain 36 (WDR36/GLC1G).15 Each of these genes is responsible for a small proportion of POAG cases (1–5%). In addition, heterozygous loss of function mutations in CYP1B1 constitutes a risk factor for development of POAG.12,16–18 Previous GWAS studies of advanced POAG identified two loci at TMCO1 and CDKN2B-AS1.19 In addition, non-advanced POAG studies have implicated CAV1,20 SIX6, and a region at 8q22.11 Recently, a three-stage GWAS identified additional genetic loci associated with POAG in participant of European descent population. It identified three new loci associated with development of POAG; these loci are located upstream of ABCA1, within AFAP1, and within GMDS. All these genes are expressed within human retina, optic nerve, and trabecular meshwork.21
These studies used different populations with different strategies, and all identified the same gene (ABCA1) which is involved in the regulation of cellular cholesterol, lipid metabolism, and playing a role in the eye disease. ABC subfamily are found exclusively in multicellular eukaryotes. ABCA1 gene is mapped to position 9q31.1. It is highly expressed in human retina, optic nerve, trabecular meshwork, and retinal ganglion cells which have a role in the development of glaucoma.21
The aim of this study was to investigate the allelic and genotypic distribution of SNPs in the ABCA1 gene in Jordanian patients of Arab descent diagnosed with primary glaucoma and to investigate the association between the ABCA1 gene polymorphisms and glaucoma among Jordanian patients of Arab descent.
Materials And Methods
Sample Study
All subjects who participated in this case-control study were of Arab descent from North Jordan including 52 unrelated patients with primary glaucoma disease and 96 healthy individuals. Clinical and demographic data were extracted and collected from their medical records. The study was approved by the Institutional Review Board (IRB) (IRB# 23/94/2016) of King Abdullah University Hospital (KAUH), Jordan. All control subjects were voluntarily involved and signed a written informed consent. All clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki consent. This study was also conducted according to the provisions of the Human Ethics standard of Jordan University of Science and Technology. Blood samples of primary glaucoma patients were collected from King Abdullah University Hospital (KAUH). The characteristics of the 52 glaucoma patients in this study were displayed in Table 1.
 |
Table 1 Characteristics Of 52 Glaucoma Patients Of Jordanian Arab Descent In This Study |
Genomic DNA Extraction
Blood samples from control and glaucoma patients were drowned into EDTA tubes. Then, genomic DNA was extracted using the commercially available kit (Qiagen, Germany) according to the manufacturer’s instructions. Briefly, 900 μL of RBC lysis solution was mixed with 300 μL of blood sample. Then, the mix was centrifuged at 16,000 rpm. 300 μL of cell lysis solution was then added to the pellet, followed by addition of 100 μL protein precipitation solution. Then, 300 μL isopropanol was added after centrifugation. The DNA was washed with 70% ethanol and dried at room temperature. Finally, 50 μL DNA rehydration solutions were added. DNA quantity and purity was verified (Bio-Drop Spectrophotometer, Cambridge, UK). The quality of the DNA samples was also evaluated on 2% agarose gel electrophoresis using the UV-light imager (Protein Simple, USA).
DNA Sequencing
PCR primers were designed to cover the coding sequences, plus a few nucleotides in the intron on both ends (Table 2). Primer extension sequencing was performed by GENEWIZ, Inc. (South Plainfield, NJ, USA) using Applied Biosystems Big Dye version 3.1. Both forward and reverse strands were sequenced. The reactions were run on Applied Biosystem’s 3730×L DNA Analyzer.
 |
Table 2 The SNPs, SNPs Position And Primers Sequences For ABCA1 Gene |
Sequencing Analyses
The sequencing data were analysed with Lasergene SeqMan (DNASTAR, Madison, WI, USA). The resulting sequence were analysed by alignment of the standard reference sequence of each primer product and then comparing the resulted genotypes to the standard ABCA1 gene reference sequence.
Genotype Phenotype Association With Clinical Variable Data Of Glaucoma Patients
A group of statistical tests were conducted to show the association between genotype and phenotype of the two reported SNPs (rs2472493 and rs2487032) in the ABCA1 gene by adjusting the clinical variable data including age, gender, family history, IOP, optic disc status, slit lamp biomicroscopy, gonioscopy, and automated visual field test in glaucoma patients. The schematic structure of the human ABCA1 and the positions of the two SNPs genotyped in this study and their dbSNP IDs are also shown in Figure 3.
Statistical Analyses
Data analysis was performed using SAS (Statistical Analysis System, version 9.2; SAS Institute, Cary, NC, USA). The genotype frequency was calculated and tested by using the Hardy-Weinberg Equilibrium equation (HWE). The allelic and genotypic association p-value was determined using Chi-squared test between cases and controls. Odds ratios were computed for each model (codominant, dominant, recessive, and overdominant) separately using SAS functions and SNPstats software (https://www.snpstats.net). The level of significance was considered at α = 0.05.
Results
Sample Characteristics
The study involved 52 Jordanian Arab patients with primary glaucoma disease and 96 healthy controls. A total of 52 Jordanian primary glaucoma patients fulfilled the exclusion criteria and consented to participate in this study. The frequency of allele and genotype for both SNPs in glaucoma patients and controls (Case/Control) are summarized in Table 3. Among 52 primary glaucoma patients, about 39 (75%) of them were diagnosed with POAG with age >35, while 13 (25%) of the patients were diagnosed with PCG with age ≤35 years. The males consist about 67.31% which compromise the large percent, while females consist only 32.69% of all patients. In the present study, the 52 Jordanian patients were diagnosed with primary glaucoma based on optic disc damage and visual field defects. IOP was more than 21 mmHg in 21 patients although some of them have been subjected to treatment. Patients with secondary causes of glaucoma (e.g. uveitis, steroid-induced, or trauma) were excluded from this study. We recruited 96 healthy controls and all were found to have no signs or symptoms of primary glaucoma or any other eye disease and did not have any family history of glaucoma.
 |
Table 3 The Frequency Of Allele And Genotype For Both SNPs In Glaucoma Patients And Controls (case/control) |
DNA Sequencing Results
SNPs Genotyping And Variation Detection Within ABCA1 Gene
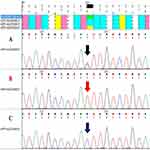
Two Single Nucleotide Polymorphisms SNPs were genotyped in order to check their presence in 52 Jordanian glaucoma patients. The resultant sequences (Electropherograms) were blasted against the reference sequence. Panel A shows a heterozygous rs2472493 sequence variant genotype, which shows both nucleotide T (red peak) and nucleotide C (blue peak); meanwhile, panel B shows the wild-type allele (T) of rs2472493 SNP for ABCA1 gene. Moreover, panel C shows homozygous rs2472493 sequence variant genotype exhibiting the conversion of nucleotide T (red peak) to nucleotide C (blue peak) (Figure 1). The Partial Electropherogram sequences of rs2487032 SNP for ABCA1 gene in glaucoma patients were shown (Figure 2). Briefly, Forward primer was used here in PCR sequencing step. Panel A shows homozygous rs2487032 sequence variant genotype exhibiting the conversion of nucleotide T (red peak) to nucleotide C (blue peak); meanwhile, panel B shows a heterozygous rs2487032 sequence variant genotype exhibiting both nucleotide T (red peak) and nucleotide C (blue peak). Finally, panel C shows the wild-type allele (T) of rs2487032 SNP for ABCA1 gene.
 |
Figure 3 Schematic structure of the human ABCA1 gene. The positions of the two SNPs genotyped and their dbSNP IDs are also shown. |
Allelic And Genotyping Distribution Of Glaucoma Patients And Healthy Controls
Genotypic and allelic frequencies were estimated after analysing ABCA1 SNPs. Table 4 shows the genetic association of two SNPs in the ABCA1 gene in glaucoma patients by age adjustment. A group of biostatistical analysis was conducted to determine if the genotype frequency was significant. However, no significant differences were observed in the rs2472493 genotype and allele frequencies between glaucoma cases and controls (P = 0.71 for genotype and P = 0.696 for allele). In addition, no significant differences were observed in the rs2487032 genotype and allele frequencies between glaucoma cases and controls (P = 0.54 for genotype and P = 0.538 for allele). Genetic association analysis of the two SNPs genotyped in the glaucoma cases and controls was also conducted using dominant, additive, and recessive genetic models. For rs2472493, the common homozygous TT (P = 0.93), common heterozygous TC (P = 0.46), and rare homozygous CC (P = 0.51) were statistically significant (P < 0.05). In contrast, for the other SNP, rs2487032, the common homozygous TT (P = 0.42), common heterozygous TC (P = 0.31), and rare homozygous CC (P = 0.92) were statistically significant (P < 0.05). Moreover, Table 5 shows the genes which play an important role in primary open angle glaucoma. The table compares the patient’s results with the controls in order to find a significant relationship between the variants and the glaucoma disease. The significance was determined according to the P-value; the level of significance taken as p < 0.05.
 |
Table 4 Genetic Association Of Two SNPs In The ABCA1 Gene In Glaucoma Patients By Age Adjustment |
 |
Table 5 Genes, Which Play Important Role In Primary Open Angle Glaucoma |
Discussion
Glaucoma is a complex disorder affected by multiple genetic and environmental factors.22 Recent GWAS have focused on identifying genes associated with glaucoma. GWAS have been very effective in mapping disease susceptibility genes. In addition, susceptibility loci for glaucoma have been mapped in many different populations; some of which have been observed in multiple populations and some of which are unique to a specific population. To date, there is a lack of GWAS performed on Middle Eastern populations which gives little opportunity to elucidate more about the aetiology of common disease in these populations. Several genetic studies including candidate genes, linkage analysis studies, and GWAS approaches have identified several loci associated with development of POAG.7,11 Recently, a three-stage GWAS identify additional genetic loci associated with POAG in participant of European descent. These loci are located upstream of ABCA1, mainly within AFAP1 and GMDS genes. All these genes are expressed within human retina, optic nerve, and trabecular meshwork.21 These studies were conducted using different populations with different strategies, and all identified the same gene (ABCA1), which is involved in the regulation of cellular cholesterol, lipid metabolism, and playing a role in the eye disease.23
Furthermore, ABCA1 gene is highly and specifically expressed in retinal ganglion cells having a role in the development of glaucoma.24 To the best of our knowledge, this study is the first in the field of ophthalmic genetics research in Jordan, and in Arab descent in general. It shows the genetic association analysis for ABCA1 gene between 52 Jordanian patients with glaucoma and 96 healthy individuals. This cohort (N=148) was used to investigate the genetic association between variants in ABCA1 gene and glaucoma risk. Previously, (rs2472493 and rs2487032) SNPs located upstream of the ABCA1 gene on 9q31.1 were identified for being associated with POAG.24–26 GWAS and meta-analysis of 18 population cohorts have discovered that rs2472493 was associated with POAG and elevated IOP.26 Meanwhile, Gharahkhani et al25 discovered similar results in Australia.
In addition, a significant genome-wide association of POAG (1007 cases) with elevated IOP (1009 controls) has been observed at multiple SNPs near ABCA1 gene on 9q31.1 using subjects from southern China.24 Multiple population replications indicated that ABCA1 gene might play a very important role in the process of POAG. On the other hand, a case-control study of the (rs2472493 and rs2487032) SNPs near the ABCA1gene among 1122 PACG subjects and 1311 healthy controls in a Han Chinese cohort showed no significant association in the distribution of allele frequencies in PACG,8 although they showed highly significant association in POAG.24–26 Even though both PACG and POAG are subtypes of glaucoma, they share many similar clinical characteristics, but the genetic susceptibility might be different on the same candidate gene.8 In this study, we genotyped two SNPs located upstream of ABCA1 gene (rs2472493 and rs2487032) in a Jordanian Arab glaucoma patients. As a result, the two reported SNPs showed no significant association in the distribution of the allele frequencies. On the contrary, they showed highly significant association in POAG from other ethnicities.24–26 This could be due to the ethnic differences between populations which might have contributed to the genetic variation. This study included only Jordanian population which is considered as an isolated population. Moreover, the genetic heterogeneity of glaucoma disorder and the possibility of the involvement of other genes or environmental factors such as gene–gene interaction or gene–environmental interaction must be considered in further research.
Conclusion
In conclusion, this study is the first genetic analysis of the ABCA1 gene in Jordanian glaucoma patients. No previous studies have been conducted on ABCA1 and its association with glaucoma in Arab nations. Herein, we relied on a sensitive sequencing technique for the genotyping process. The findings of this study proved that no association was observed between these SNPs and glaucoma in Jordanian Arab descent. This suggests that the genetic background of glaucoma in Jordanian Arab ethnicity and other ethnicities are different. Thus, further research with large sample size is needed to elucidate the exact mechanism of ABCA1 in the progress of glaucoma in Jordan and Arab nation populations.
Acknowledgments
This work was financially supported by the Deanship of Scientific Research at Jordan University of Science and Technology for their generous grant No. 2016/107. The authors also thank the Ophthalmic Clinic in King Abdullah University Hospital (KAUH) for their help in sample collection.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Quigley H, Addicks E. Regional differences in the structure of the lamina cribrosa and their relation to glaucomatous optic nerve damage. Arch Ophthalmol. 1981;99:137–143. doi:10.1001/archopht.1981.03930010139020
2. Kielczewski J, Pease M, Quigley H. The effect of experimental glaucoma and optic nerve transection on amacrine cells in the rat retina. Invest Ophthalmol Vis Sci. 2005;46:3188–3196. doi:10.1167/iovs.05-0321
3. Tatjana C, Jakobs RT, Ben LY, John SWM, Masland RH. Retinal ganglion cell degeneration is topological but not cell type specific in DBA/2J mice. J Cell Biol. 2005;171:313–325. doi:10.1083/jcb.200506099
4. Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262–267. doi:10.1136/bjo.2005.081224
5. Allingham RR, Damji KF, Shields MB. Shields Textbook of Glaucoma.
6. Racette L, Wilson MR, Zangwill LM, Weinreb RN, Sample PA. Primary open-angle glaucoma in blacks: a review. Surv Ophthalmol. 2003;48:295–313. doi:10.1016/S0039-6257(03)00028-6
7. Stone EM, Fingert JH, Alward WL, et al. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–670. doi:10.1126/science.275.5300.668
8. Luo H, Chen Y, Ye Z, et al. Evaluation of the association between common genetic variants near the ABCA1 gene and primary angle closure glaucoma in a Han Chinese population. Invest Ophthalmol Vis Sci. 2015;56:6248–6254. doi:10.1167/iovs.15-16741
9. Kumar A, Basavaraj MG, Gupta SK, Qamar I, Ali AM, Bajaj V. Role of CYP1B1, MYOC, OPTN and OPTC genes in adult-onset primary open-angle glaucoma: predominance of CYP1B1 mutations in Indian patients. Mol Vis. 2007;13:667–676.
10. James B, Born A. Ophthalmology Lecture Notes.
11. Wiggs JL, Yaspan BL, Hauser MA, Kang JH, Allingham RR, Olson LM. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet. 2012;e1002654. doi:10.1371/journal.pgen.1002654
12. Chakrabarti S, Devi KR, Komatireddy S, et al. Glaucoma-associated CYP1B1 mutations share similar haplotype backgrounds in POAG and PACG phenotypes. Invest Ophthalmol Vis Sci. 2007;48:5439–5444. doi:10.1167/iovs.07-0629
13. Fan BJ, Wang DY, Lam DS, Pang CP. Gene mapping for primary open angle glaucoma. Clin Biochem. 2006;39:249–258. doi:10.1016/j.clinbiochem.2005.10.013
14. Rezaie T, Child A, Hitchings R. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295:1077–1079. doi:10.1126/science.1066901
15. Monemi S, Spaeth G, DaSilva A, Popinchalk S, Ilitchev E, Liebmann J. Identification of a novel adult-onset primary open angle glaucoma (POAG) gene on 5q22.1. Hum Mol Genet. 2005;14:725–733. doi:10.1093/hmg/ddi068
16. Melki R, Colomb E, Lefort N, Brézin AP, Garchon H-J. CYP1B1 mutations in French patients with early-onset primary open-angle glaucoma. J Med Genet. 2004;41:647–651. doi:10.1136/jmg.2004.020024
17. Acharya M, Mookherjee S, Bhattacharjee A, Bandyopadhyay AK, Thakur SK, Bhaduri G. Primary role of CYP1B1 in Indian juvenile-onset POAG patients. Mol Vis. 2006;12:399–404.
18. Pasutto F, Chavarria-Soley G, Mardin CY. Heterozygous loss of function variants in CYP1B1 predispose to primary open angle glaucoma. Invest Ophthalmol Vis Sci. 2009;51:249–254.
19. Burdon KP, Macgregor S, Hewitt AW, et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B–AS1. Nat Genet. 2011;43:574–578. doi:10.1038/ng.824
20. Thorleifsson G, Walters GB, Hewitt AW. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet. 2010;42:906–909. doi:10.1038/ng.661
21. Craig JE. Common variants near ABCA1, AFAP1, and GMDS confer risk of primary open angle glaucoma. Nat Genet. 2014;46:1120–1125. doi:10.1038/ng.2895
22. Quigley HA. Number of people with glaucoma worldwide. Br J Ophthalmol. 1996;80:389–393. doi:10.1136/bjo.80.5.389
23. Santamarina-Fojo S, Peterson K, Knapper C, et al. Complete genomic sequence of the human ABCA1 gene: analysis of the human and mouse ATP-binding cassette A promoter. Proc Natl Acad Sci USA. 2000;97:7987–7992. doi:10.1073/pnas.97.14.7987
24. Chen Y, Lin Y, Vithana EN, et al. Common variants near ABCA1 and in PMM2 are associated with primary open-angle glaucoma. Nat Genet. 2014;46:1115–1119. doi:10.1038/ng.2895
25. Gharahkhani P, Burdon KP, Fogarty R, Sharma S, Hewitt AW, Martin S. Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat Genet. 2014;46:1120–1125. doi:10.1038/ng.2895
26. Hysi PG, Cheng CY, Springelkamp H, Macgregor S, Bailey JN, Wojciechowski R. Genome-wide analysis of multi-ancestry cohorts identifies new loci influencing intraocular pressure and susceptibility to glaucoma. Nat Genet. 2014;46:1126–1130. doi:10.1038/ng.2895
27. Osman W, Low SK, Takahashi A, Kubo M, Nakamura YA. Genome-wide association study in the Japanese population confirms 9p21 and 14q23 as susceptibility loci for primary open angle glaucoma. Hum. Mol. Genet. 2012;21:2836–2842.
28. Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K, Saito K, Yonemura S, Eiraku M, Sasai Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.