Back to Journals » OncoTargets and Therapy » Volume 8
Gefitinib upregulates death receptor 5 expression to mediate rmhTRAIL-induced apoptosis in Gefitinib-sensitive NSCLC cell line
Authors Yan D, Ge Y, Deng H, Chen W, An G
Received 4 September 2014
Accepted for publication 4 May 2015
Published 3 July 2015 Volume 2015:8 Pages 1603—1610
DOI https://doi.org/10.2147/OTT.S73731
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jianmin Xu
Dong Yan,1,2 Yang Ge,1 Haiteng Deng,3 Wenming Chen,4 Guangyu An1
1Department of Oncology, Beijing Chao-yang Hospital, Capital Medical University, Beijing, People’s Republic of China; 2Translational Molecular pathology, M.D Anderson Cancer Center, Houston, TX, USA; 3School of Sciences, Tsinghua University, 4Department of Hematology, Beijing Chao-yang Hospital, Capital Medical University, Beijing, People’s Republic of China
Background: Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) triggers apoptosis in tumor cells, but when used alone, it is not effective in the treatment of TRAIL-resistant tumors. Some studies have shown that gefitinib interacts with recombinant mutant human TRAIL (rmhTRAIL) to induce high levels of apoptosis in gefitinib-responsive bladder cancer cell lines; however, the molecular mechanisms underlying the anticancer effects are not fully understood. Several reports have shown that the death receptor 5 (DR5) plays an important role in sensitizing cancer cells to apoptosis induced by TRAIL. Therefore, we investigated the effects of the combination of drugs and the expression of the DR5 to analyze the growth of a gefitinib-responsive non-small cell lung cancer cell line PC9, which was treated with rmhTRAIL and gefitinib individually or in combination.
Methods: Human PC9 non-small cell lung cancer cells harboring an epidermal growth factor receptor mutation were used as a model for the identification of the therapeutic effects of gefitinib alone or in combination with rmhTRAIL, and cytotoxicity was assessed by MTT assays. Cell cycle and apoptosis were investigated using flow cytometry. Moreover, the effects of drugs on DR5, BAX, FLIP, and cleaved-caspase3 proteins expressions were analyzed using Western blot analyses. Finally, quantitative polymerase chain reaction analysis was carried out to assess whether rmhTRAIL and gefitinib modulate the expression of genes related to drug activity.
Results: Gefitinib and rmhTRAIL synergistically interact to inhibit cell proliferation, and apoptosis assessment demonstrated that associations of drug increased the apoptotic index. rmhTRAIL when used alone downregulated DR5 and upregulated BAX, FLIP, and cleaved-caspase3 proteins expressions. However, results obtained in Western blot analyses demonstrated that the combined treatment-induced cell apoptosis was achieved involving upregulated DR5, cleaved-caspase3, and BAX proteins expression and downregulated FLIP protein expression. Moreover, quantitative polymerase chain reaction showed that gefitinib modulated the expression of targets related to rmhTRAIL activity.
Conclusion: These results indicate that epidermal growth factor receptor inhibitors enhance rmhTRAIL antitumor activity in the gefitinib-responsive PC9 cell line, and upregulated DR5 expression plays a critical role in activating caspase-signaling apoptotic pathway.
Keywords: gefitinib, rmhTRAIL, apoptosis, DR5
Introduction
Non-small cell lung cancer (NSCLC) is the leading cause of death due to cancer worldwide. Despite improvements in chemoradiotherapy and targeted therapy, the long-term survival of patients with NSCLC remains low. Therefore, novel and more effective treatments and strategies for NSCLC are critically needed.
Apoptosis is executed through two main pathways (the extrinsic and intrinsic pathways), and both these pathways can be targeted therapeutically.1,2 Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) belongs to a group of proapoptotic cytokines in the tumor necrosis factor superfamily.3 TRAIL is known to bind to death receptors such as death receptor 5 (DR5) and/or death receptor 4, and this binding allows for the formation of a death-inducing signaling complex (DISC), resulting in the activation of caspase-signaling pathways leading to apoptosis.4 However, tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) triggers apoptosis in tumor cells, but when used alone, it is ineffective for the treatment of TRAIL-resistant tumors.
Epidermal growth factor receptor–tyrosine kinase inhibitor (gefitinib) is a targeted agent that has been initially approved for the treatment of patients with advanced or metastatic NSCLC who have epidermal growth factor receptor (EGFR) mutations.5,6 Gefitinib blocks the intracellular ATP-binding site of EGFR and thereby interrupts downstream signal transduction. Some studies have shown that gefitinib interacts with recombinant mutant human TRAIL (rmhTRAIL) to induce high levels of apoptosis in gefitinib-responsive bladder cancer cell lines.7,8 However, the molecular mechanisms underlying the anticancer effects of gefitinib on TRAIL-induced apoptosis are not fully understood. Several reports have shown that the DR5 plays an important role in sensitizing cancer cells to apoptosis induced by TRAIL.9,10 Therefore, we used human NSCLC PC9 cells harboring an EGFR mutation as a model to identify whether the therapeutic combinations of EGFR inhibitor with rmhTRAIL dramatically induce apoptosis-dependent DR5 upregulation.
Methods
Reagents
Gefitinib and rmhTRAIL freeze-dried powders obtained from AstraZeneca (Beijing, People’s Republic of China) and Shadong pharmaceuticals (Beijing, People’s Republic of China) were dissolved in dimethyl sulfoxide and distilled water, respectively, at stock concentrations of 105 mol/L and 106 ng/mL and diluted to indicated concentrations with Roswell Park Memorial Institute medium, (RPMI)-1640 medium. RPMI-1640 medium was purchased from Gibco (Invitrogen, Life Technologies, Shanghai, People’s Republic of China). Trypsin and MTT were purchased from Sigma (Shanghai, People’s Republic of China). Antibodies against BAX, cleaved-caspase3, FLIP, and DR5, were from Cell Signaling Technology Inc.
Cell culture and research methods
Human lung cancer A549 (adenocarcinoma), PC9 (adenocarcinoma), and H358 (adenocarcinoma) cells were obtained from Beijing institute for cancer research. Cells were grown in RPMI with 10% fetal bovine serum and 1% penicillin–streptomycin. Cells were cultured in 75 cm2 flasks at 37°C in 5% CO2 and 95% air.
Cytotoxicity assay
Cells were plated in six-well sterile plastic plates at 105 cells/well to facilitate the attachment to the substrate for 24 hours. PC9 cells were treated with (a) gefitinib (0.01–1 μmol/L) for 72 hours followed by a 24-hour washout in drug-free medium, (b) rmhTRAIL (0.1–1,000 ng/mL) for 72 hours, and (c) rmhTRAIL (50 ng/mL) and gefitinib (0.01 μmol/L) for 72 hours, and PBS served as a blank control. At the end of drug exposure, 15 μL MTT solution was added, and cells were incubated in a humidified 5% CO2 atmosphere. After 4 hours, a solubilization solution was added to allow the full solubilization of formazan crystals. Cell growth inhibition was expressed as a percentage of the absorbance of control cultures measured at 630 nm using a microplate reader (Multiskan MK3; Thermo, USA), and the 50% inhibitory concentration of cell growth (IC50) was calculated by GraphPad Prism. All experiments were repeated three times. Drug interaction between gefitinib and rmhTRAIL were assessed using the combination index (CI), where CI<1, CI=1, and CI>1 indicate synergistic, additive, and antagonistic effects, respectively.11
Cell cycle and apoptosis analyses
Cell cycle distribution was determined by measuring the cellular DNA content using flow cytometry. Cells were trypsinized, washed in PBS, fixed in 70% ice-cold ethanol at 4°C overnight, washed again with PBS, and stained with 100 mL of 50 mg/L propidium iodide at 37°C for 30 minutes. An annexin V-FITC apoptosis detection kit was used to identify apoptotic and viable cells following the manufacturer’s instructions. The percentage of early apoptotic (FITC-positive and propidium iodide-negative) cells was calculated from the data generated by flow cytometry.
Western blot analyses
Lysates were separated on 12.5% sodium dodecyl sulfate gels before transferring to polyvinylidene difluoride membranes. The membranes were probed with primary antibodies (DR5, BAX, FLIP, and cleaved-caspase3 at 1:500), washed with Tris-buffered saline with Tween (TBST), followed by secondary antibodies (at 1:5,000), and conjugated with horseradish peroxidase. Immunoreactivities were detected using an ECL Plus™ kit, and quantitative data were obtained using molecular imaging software. Three independent experiments were performed.
Quantitative PCR
Total RNA was extracted using the RNeasy kit (Qiagen). Two micrograms of RNA from each sample was used as template for cDNA synthesis with a reverse transcription kit (Promega). An equal volume of cDNA product was used for polymerase chain reaction (PCR). The primers used for PCR were as follows: DR5 (199bp), 5′–GGGATGGTCAAGGTCGGTG–3′ (forward) and 5′–CAGCCACAATCAAGACTACGG–3′ (reverse); caspase3 (165 bp), 5′–AGAACTG GACTGTGGCATTGAG–3′ (forward) and 5′–GCACAAAGCGACTGGATGAA–3′ (reverse); actin (205 bp), 5′–TGACGTGGACATCCGCAAAG3′ (forward) and 5′–CTGGAAGGTGGACAGCGAGG–3′ (reverse). The quantitative PCR (qPCR) reactions were run with an FTC2000 sequence detection system (initial incubation at 95°C for 4 minutes followed by 40 cycles of 95°C for 20 seconds, 60°C for 30 seconds, and 72°C for 30 seconds for extension), and the resulting data were analyzed using Sequence Detection System software. Samples were assigned a Ct value for each gene (the cycle number at which the logarithmic PCR plots cross a fixed threshold).
Mutation analysis
PCR products were directly sequenced to reveal possible mutations. Samples identified as wild type were used as negative controls, and samples mutated with a 746–751 deletion in exon 19 or an L858R mutation in exon 21 of the EGFR gene, were used as positive controls. Water was used as a blank control. The KRAS gene harbors two hot spots for activating mutations (KRASG12/13).
Statistical analysis
All experiments were repeated independently for a minimum of three times and expressed as mean values with 95% confidence intervals. All statistical calculations were performed using SPSS software and GraphPad Prizm 5.0. Statistical analysis was carried out using the chi-square test, Fisher’s exact test, nonparametric analysis of variance (ANOVA), and independent-samples t-test. Differences were considered significant when P≤0.05.
Results
Molecular characteristics and cytotoxicity of gefitinib and rmhTRAIL in human NSCLC cell line PC9
The status of EGFR and KRAS gene mutation of human NSCLC cells including A549, H358, and PC9 were examined. Only PC9 cells harbor an in-frame deletion in exon 19 of EGFR and were found to be highly sensitive to gefitinib.12 To confirm KRAS mutation in NSCLC cell lines, direct DNA sequencing of KRAS exon 2 was performed. Wild-type sequence was detected in PC9, whereas others cells had a point mutation (Figure 1). Our study showed the cytotoxic effect in human PC9 cell line by gefitinib or rmhTRAIL in a concentration- and time-dependent manner. Gefitinib was cytotoxic against PC9 cells with IC50s of 0.02±0.01 μmol/L. Cell growth inhibition was also observed after 72 hours of exposure to rmhTRAIL, with IC50s of 84.9±9.86 ng/mL in PC9 cells (Figure 2). As had been found previously for TRAIL receptor targeting therapies in NSCLC, PC9 cells were considered nonsensitive to rmhTRAIL.8
Assay of interaction between gefitinib and rmhTRAIL
As human PC9 NSCLC cells that carry an EGFR mutation were shown to be very sensitive to gefitinib and nonsensitive to rmhTRAIL, this cell line was selected as a model to evaluate the cytotoxic interaction between gefitinib and rmhTRAIL. Since the CI method recommends a ratio of IC50 values at which drugs are equipotent, combination studies were carried out at different doses (0.005 μmol/L, 0.01 μmol/L, 0.05 μmol/L, 0.1 μmol/L, 0.5 μmol/L) of gefitinib and rmhTRAIL (50 ng/mL), whereas for individual studies, doses (5 ng/mL, 10 ng/mL, 25 ng/mL, 50 ng/mL, 125 ng/mL) of rmhTRAIL and gefitinib (0.01 μmol/L) at different time points (24 hours, 48 hours, 72 hours) were studied. The growth inhibition increased when rmhTRAIL was combined with gefitinib at a certain dose for 48 hours–72 hours (Figure 2). The analysis of drug interaction revealed synergistic effects (CI<1) at 50% effect level on cell growth inhibition in combination treatments (Table 1).
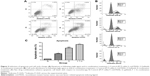
Induction of apoptosis and cell cycle change
Apoptosis and cell cycle distribution were analyzed using flow cytometry after single agent and concurrent drug treatment for 48 hours. Upon exposure to gefitinib, rmhTRAIL, and their combinations, lung cancer cells presented typical apoptotic morphology with cell shrinkage, nuclear fragmentation, and cellular rupture into debris. The occurrence of apoptosis was significantly higher in cells treated with gefitinib and rmhTRAIL as single agents with respect to controls (Figure 3). Moreover, the combination of the two drugs enhanced apoptotic cell death with respect to monotherapy in PC9 cell lines (Table 2). rmhTRAIL, at IC50 levels, enhanced cellular population with sub-G1 DNA content with respect to controls, whereas after 48 hours of treatment with gefitinib, G1 cell cycle arrest was induced. However, the appearance of a sub-G1 peak was significantly increased by rmhTRAIL and gefitinib (Figure 3).
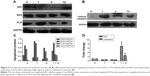
Combined gefitinib and rmhTRAIL treatment mediates the expression of various apoptotic proteins
To investigate the role and mechanism by which gefitinib enhanced TRAIL-induced apoptosis, the level of apoptosis signaling molecules, such as DR5, BAX, FLIP, and cleaved-caspase3 proteins, of the extrinsic or death receptor pathway was evaluated. Our results suggest that EGFR inhibitors sensitized rmhTRAIL antitumor activity in NSCLC cell line via upregulation of DR5, cleaved-caspase 3, and BAX expression, and downregulation of FLIP protein expression (Figure 4).
Modulation of gene expression of DR5 and caspase3 genes by gefitinib and rmhTRAIL
Gefitinib and rmhTRAIL downregulated DR5 gene expression in PC9 cells. A significantly enhanced gene expression of DR5 and caspase3 was observed in PC9 cells after treatment with gefitinib and rmhTRAIL (Figure 4).
Discussion
EGFR inhibitors have been important therapies for NSCLC. Studies have shown that EGFR-TKIs, such as gefitinib, can produce objective response and prolong progression-free survival in patients with NSCLC with EGFR mutation.13–15 TRAIL therapy is effective against many cancer types, including NSCLC; yet, in some NSCLC cell lines, the effect is limited.16 Inherent tumor resistance may be a major barrier to effective TRAIL-targeted therapy,17 indicating that sensitization of cancer cells to the TRAIL-induced apoptotic pathway is likely to be an important anticancer strategy. However, it is unclear whether gefitinib enhances TRAIL-induced apoptosis of NSCLC cells. In the present study, our results show that rmhTRAIL had a dose-dependent cytotoxic effect in NSCLC PC9 cell line, and this effect became significant at high MOL. Furthermore, rmhTRAIL in combination with gefitinib significantly suppressed the growth of gefitinib-sensitive PC9 cells, suggesting that gefitinib enhances sensitivity of cancer cells to the apoptotic agent TRAIL and enhances its potential use as a novel strategy to current cancer therapies.
TRAIL induces death of malignant cells or apoptosis via the extrinsic apoptotic pathway, and the EGFR pathway and extrinsic apoptosis pathway interact closely in epithelial-derived cells.18 Some studies showed that EGF protects epithelial-derived cells from TRAIL-induced apoptosis.19 Therefore, inhibition of the EGFR pathway may enhance TRAIL-induced apoptosis. However, the basis of synergistic interaction between gefitinib and rmhTRAIL remains unclear. In the present work, we used human NSCLC cell line PC9 that harbors an EGFR mutation as a gefitinib-sensitive model to examine the effect of events involved in the inhibition of cell proliferation and cell death induced by gefitinib, rmhTRAIL, or the combination of both. Proliferation of PC9 cell line was inhibited when low concentration of rmhTRAIL was combined with EGFR-TKIs, at which the inhibitors had light-to-mild cytotoxic effects. We selected the PC9 cell line, which was responsive to EGFR inhibitors and had obvious loss of cell viability after combination treatment, to determine whether this synergism was dependent on the enhancement of rmhTRAIL activity. As shown in our study, gefitinib and rmhTRAIL caused a time- and concentration-dependent inhibition of cell growth as single agents in PC9 lung cancer cells. The exposure of PC9 cells to combination regimes results in synergistic effect in the inhibition of cell proliferation and apoptotic cell death as verified by the results obtained from MTT assay and the assessment of sub-G1 cells by flow cytometry.
However, the molecular mechanisms underlying the anticancer effects of gefitinib on TRAIL-induced apoptosis are not fully understood. The binding of TRAIL to TRAIL-R1 (DR4) or TRAIL-R2 (DR5) results in trimerization of the receptors and formation of a DISC consisting of Fas-associated death domain protein and procaspase-8 that is then cleaved and activated. The protein c-FLIP can also be used in the DISC thereby preventing caspase-8 binding and activation leading to suppression of apoptosis.20,21 The extrinsic apoptotic pathway initiated by caspase-8 activation will be sufficient for irreversible apoptosis activation directly through caspase3 activation, although often cross-activation of the intrinsic route via caspase-8-dependent Bid cleavage is required, as is the case for NSCLC cells.7 Our results showed that rmhTRAIL when used alone caused significant decrease in the levels of DR5 protein and increased c-FLIP protein expression in NSCLC PC9 cells. It is suggested that the downregulation of DR5 and upregulation of c-FLIP were resistant to cell death for PC9 cells exposed to rmhTRAIL. Interestingly, combination treatment can increase the levels of cleaved-caspase3 and downregulate FLIP expression, emphasizing the specific effect of upregulation of DR5 protein and gene expression compared with rmhTRAIL when used alone. The increased level of DR5 expression, leading to stimulation of the death receptor pathway, seems to be the cause of the activation of caspase3. Therefore, enhanced TRAIL-induced apoptosis by gefitinib-induced DR5 upregulation is associated with increased activation of the caspase cascade and apoptosis. In addition, in accordance with the previous reports demonstrating the relationship between gefitinib-induced mitochondrial dysfunction and cell apoptosis,22 we also showed that gefitinib increased the mitochondrial apoptosis proteins such as BAX, leading to the enhanced apoptosis of cancer cells to TRAIL.
Conclusion
In summary, our results demonstrate that combination of rmhTRAIL and gefitinib could improve sensitivity to rmhTRAIL in gefitinib-sensitive NSCLC cells by upregulation of DR5, BAX, and cleaved-caspase3 expression and downregulated c-FLIP expression involved in extrinsic and intrinsic apoptotic pathways. However, in vitro drug trials may be different from that of in vivo given the impact of human pharmacokinetics. Usually, drug tests are more complex in vivo. For this reason, our findings may need further research to clarify the molecular mechanisms underlying the present important results, but this research provided us with a promising treatment that will bring hope to the clinical treatment of patients with NSCLC.
Acknowledgments
We thank Professor Wenming Chen, for her constant encouragement and guidance. The study has received funding from Technology project of Beijing traditional Chinese medicine (JJ2013-18).
Disclosure
The authors declare that they have no competing interests.
References
Ashkenazi A, Herbst RS. To kill a tumor cell: the potential of proapoptotic receptor agonists. J Clin Invest. 2008;118:1979–1990. | ||
Jin Z, El-Deiry WS. Overview of cell death signaling pathways. Cancer Biol Ther. 2005;4(2):139–163. | ||
Oikonomou E, Pintzas A. The TRAIL of oncogenes to apoptosis. Biofactors. 2013;39(4):343–354. | ||
Minakawa T, Toume K, Arai MA, Koyano T, Kowithayakorn T, Ishibashi M. Prenylflavonoids isolated from Artocarpus champeden with TRAIL-resistance overcoming activity. Phytochemistry. 2013;96:299–304. | ||
Soria JC, Mok TS, Cappuzzo F, Jänne PA. EGFR-mutated oncogene-addicted non-small cell lung cancer: current trends and future prospects. Cancer Treat Rev. 2012;38(5):416–430. | ||
Reungwetwattana T, Dy GK. Targeted therapies in developent for non-small cell lung cancer. J Carcinog. 2013;12:22. | ||
Stegehuis JH, de Wilt LH, de Vries EG, Groen HJ, de Jong S, Kruyt FA. TRAIL receptor targeting therapies for non-small cell lung cancer: current status and perspectives. Drug Resist Updat. 2010;13(1–2):2–15. | ||
Shrader M, Pino MS, Lashinger L, et al. Gefitinib reverses TRAIL resistance in human bladder cancer cell lines via inhibition of AKT-mediated X-linked inhibitor of apoptosis protein expression. Cancer Res. 2007;67(4):1430–1435. | ||
Omar HA, Arafa ES, Maghrabi IA, Weng JR. Sensitization of hepatocellular carcinoma cells to Apo2L/TRAIL by a Novel Akt/NF-κB signalling inhibitor. Basic Clin Pharmacol Toxicol. 2014;114(6):464–471. | ||
Kim EH, Yoon MJ, Kim SU, Kwon TK, Sohn S, Choi KS. Arsenic trioxide sensitizes human glioma cells, but not normal astrocytes, to TRAIL-induced apoptosis via CCAAT/enhancer-binding protein homologous protein-dependent DR5 up-regulation. Cancer Res. 2008;68(1):266–275. | ||
Lou YF, Zou ZZ, Chen PJ, et al. Combination of gefitinib and DNA methylation inhibitor decitabine exerts synergistic anti-cancer activity in colon cancer cells. PLoS One. 2014;9:e97719. | ||
Fukuoka M, Yano S, Giaccone G, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial) [corrected]. J Clin Oncol. 2003;21(12):2237–2246. | ||
Ulivi P, Zoli W, Capelli L, Chiadini E, Calistri D, Amadori D. Target therapy in NSCLC patients: relevant clinical agents and tumour molecular characterization. Mol Clin Oncol. 2013;1(4):575–581. | ||
Powrózek T, Krawczyk P, Ramlau R, et al. EGFR gene mutations in patients with adenosquamous lung carcinoma. Asia Pac J Clin Oncol. 2014;10(4):340–345. | ||
Berge EM, Doebele RC. Targeted therapies in non-small cell lung cancer: emerging oncogene targets following the success of epidermal growth factor receptor. Semin Oncol. 2014;41(1):110–125. | ||
Voortman J, Resende TP, Abou EL, Hassan MA, Giaccone G, Kruyt FA. TRAIL therapy in non-small cell lung cancer cells: sensitization to death receptor-mediated apoptosis by proteasome inhibitor bortezomib. Mol Cancer Ther. 2007;6(7):2103–2112. | ||
Llobet D, Eritja N, Encinas M, et al. CK2 controls TRAIL and Fas sensitivity by regulating FLIP levels in endometrial carcinoma cells. Oncogene. 2008;27(18):2513–2524. | ||
Gibson SB. Epidermal growth factor and trail interactions in epithelial-derived cells. Vitam Horm. 2004;67:207–227. | ||
Gibson EM, Henson ES, Haney N, Villanueva J, Gibson SB. Epidermal growth factor protects epithelial-derived cells from tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by inhibiting cytochrome c release. Cancer Res. 2002;62(2):488–496. | ||
Li Z, Xu X, Bai L, Chen W, Lin Y. Epidermal growth factor receptor-mediated tissue transglutaminase overexpression couples acquired tumor necrosis factor-related apoptosis-inducing ligand resistance and migration through c-FLIP and MMP-9 proteins in lung cancer cells. J Biol Chem. 2011;286(24):21164–21172. | ||
Zang F, Wei X, Leng X, Yu M, Sun B. C-FLIP(L) contributes to TRAIL resistance in HER2-positive breast cancer. Biochem Biophys Res Commun. 2014;450:267–273. | ||
Yu SY, Liu HF, Wang SP, Chang CC, Tsai CM, Chao JI. Evidence of securin-mediated resistance to gefitinib-induced apoptosis in human cancer cells. Chem Biol Interact. 2013;203(2):412–422. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.