Back to Journals » International Journal of General Medicine » Volume 16
GBP5 Identifies Immuno-Hot Tumors and Predicts the Therapeutic Response to Immunotherapy in NSCLC
Authors Fan H, Shi Y, Wang H, Li Y, Mei J, Xu J , Liu C
Received 3 March 2023
Accepted for publication 28 April 2023
Published 10 May 2023 Volume 2023:16 Pages 1757—1769
DOI https://doi.org/10.2147/IJGM.S408900
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Honghong Fan,* Yuxin Shi,* Huiyu Wang, Yuting Li, Jie Mei, Junying Xu, Chaoying Liu
Department of Oncology, The Affiliated Wuxi People’s Hospital of Nanjing Medical University, Wuxi, Jiangsu, 214023, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chaoying Liu; Junying Xu, Department of Oncology, The Affiliated Wuxi People’s Hospital of Nanjing Medical University, No. 299 Qingyang Road, Wuxi, Jiangsu, 214023, People’s Republic of China, Tel/Fax +86-510-82700775, Email [email protected]; [email protected]
Background: Immunotherapy drugs, immune checkpoint inhibitors (ICIs), have been approved for first- and second-line treatment of non-small cell lung cancer (NSCLC), but only a portion of patients respond to ICIs. It is crucial to screen the beneficiaries of immunotherapy through biomarkers accurately.
Methods: Several datasets were used to explore the predictive value for immunotherapy and immune relevance of guanylate binding protein 5 (GBP5) in NSCLC, including the GSE126044 dataset, The Cancer Genome Atlas (TCGA) dataset, Clinical Proteomic Tumor Analysis Consortium (CPTAC) dataset, the Kaplan–Meier plotter dataset, the HLuA150CS02 cohort, and the HLugS120CS01 cohort.
Results: GBP5 was upregulated in tumor tissues but associated with a good prognosis in NSCLC. Moreover, our findings demonstrated that GBP5 was strongly correlated with the expression of many immune-related genes, TIIC levels, and PD-L1 expression based on RNA-seq data onto online databases and validation of the NSCLC tissue microarray using IHC staining. Moreover, pan-cancer analysis has shown that GBP5 was a factor in identifying immuno-hot tumors, except for a few tumor types.
Conclusion: In summary, our current research suggests that GBP5 expression is a potential biomarker for predicting the outcome of NSCLC patients treated with ICIs. More research with large-scale samples is needed to determine their value as biomarkers of ICIs benefit.
Keywords: GBP5, biomarker, immunotherapy, NSCLC, bioinformatics
Introduction
Immunotherapy for malignancies depends on the autoimmune system, which kills tumor cells and tissues by reactivating the body’s immune system. At present, immune checkpoint inhibitors (ICIs) represented by PD-1/PD-L1 antibodies have represented historic breakthroughs in the clinical immunotherapy treatment of non-small cell lung cancer (NSCLC). Immunotherapy does not have a good therapeutic effect on all NSCLC patients. Patients with different molecular, histological, or genetic characteristics have different responses to immunotherapy.1 In addition, the increased activity of the body’s immune system due to immunotherapy may also cause more terrible immune-related adverse reactions.2 Programmed cell death 1 ligand 1 (PD-L1) expression,3 tumor mutational burden (TMB),4 and microsatellite instability (MSI),5 as classic biomarkers, are not yet perfect in precisely predicting treatment effects. For example, in some large-scale clinical trials of NSCLC patients receiving immunotherapy, there was no correlation between PD-L1 expression levels with clinical treatment outcomes.6 Therefore, under the current background of precision medicine, it is the priority to eagerly search for biomarkers by comprehensive means, screen the benefit population of immunotherapy and monitor the therapeutic efficacy and prognosis.
Interferon-gamma (IFN-γ) obtains a lymphokine with antiviral biological activity and broad immunomodulatory effects. As a superfamily of GTPase proteins induced by IFN-γ activation of the JAK-STAT signaling pathway, it serves various biological functions. It is famous for its protective immunity against pathogens.7–9 Previous studies found guanylate binding protein 5 (GBP5) was highly expressed in gastric adenocarcinoma and colon cancer.10,11 In glioblastoma cells, GBP5 overexpression can promote the proliferation, migration, and invasion of glioblastoma (GBM) cells in vitro, contributing to the growth and invasion of GBM in vivo.12 Currently, immunotherapy is mainly used in clinical treatment for NSCLC patients with negative mutations in sensitive driver genes and high PD-L1 expression. Still, not all patients have a better therapeutic response to immunotherapy, so using PD-L1 expression as a predictive marker for NSCLC efficacy is not comprehensive.13 Meanwhile, Shenoy’s study found that GBP5 is associated with pyroptosis, a vital body immune response, and is crucial in tumors.14 In addition, GBP5 has been shown to enhance tumor immunogenicity and potentially be a target for immunotherapy of malignant tumors.15 However, the relevance of GBP5 as a predictive biomarker of efficacy with antitumor immunotherapy in NSCLC remains unclear.
The predictive effect of GBP5 in NSCLC patients undergoing immunotherapy was examined in this study. According to our findings, patients with NSCLC who have high GBP5 expression have a better prognosis. We discovered GBP5 expression was upregulated in NSCLC tissues compared to the para-cancer while positively related to PD-L1 expression using immunohistochemistry (IHC) analysis. GBP5 expression was immune-linked in NSCLC and most malignancies, according to the analysis of data from a cohort of NSCLC patients who received immunotherapy and The Cancer Genome Atlas (TCGA) data. These findings point to the potential predictive utility of GBP5 in determining the course of immunotherapy treatment for NSCLC patients.
Methods and Materials
Expression Analysis of GBP5 in NSCLC
After downloading the transcriptomic data as Fragments per kilobase per million (FPKM) of NSCLC from the TCGA database, we performed to explore the difference of GBP5 between tumor and para-cancer tissues. The Clinical Proteomic Tumor Analysis Consortium (CTPAC) dataset was used to analyze the expression of GBP5 protein in lung adenocarcinoma (LUAD) in normal and primary tumors.
Kaplan–Meier Plotter to Analyze the Prognosis Online
Log in to the Kaplan–Meier plotter website (http://kmplot.com/analysis/), choose the “mRNA-gene chip-lung cancer” subset for analysis, input the protein gene name GBP5, select the corresponding probe 238581_at, separate the gene expression into high and low expression groups according to their median levels, select “OS” and “PPS” for survival data type, and set all other variables by default system, then draw the Kaplan–Meier plot with p<0.05 indicating statistical significance.
Public Datasets, Data Processing, Characterization of Tumor Immune Microenvironment
A public dataset consisting of clinical and RNA-seq data from NSCLC patients undergoing immunotherapy was downloaded from the Gene Expression Omnibus (GEO) databases by following the links (http://www.ncbi.nlm.nih.gov/geo/), namely the GSE126044 dataset.The tumor immune microenvironment (TIME) characteristics contained immunomodulators, tumor purity, infiltration levels of tumor-infiltrating immune cells (TIICs), and the expression of inhibitory immune checkpoint molecules.16,17 First, we analyzed the relevance of immunomodulators to GBP5 in the TIME of NSCLC, comprising chemokines, major histocompatibility complex (MHC) molecules, receptors, immunoinhibitors, and immunostimulators. We also assessed the level of TIICs in NSCLC with two different methods: TIMER18 and EPIC19 algorithms. In addition, the relevance of GBP5 to the level of immune checkpoint expression was assessed.
For pan-cancer analysis, we also evaluated the correlation between GBP5 and immunomodulators, TIICs, TMB, tumor purity, and neoantigen mutations using Sangerbox online tool.20 The clinical annotation of the pan-cancer and NSCLC gene expression profiles was obtained from the UCSC (https://xenabrowser.net/datapages/). The abbreviations for TCGA cancer types are shown in Table S1.
Correlation Between GBP5 and Immunotherapeutic Response
Based on data from existing studies, the Calculation of immunophenoscore (IPS) values can be implemented to determine the immunogenicity of tumors and predict the response of many types of tumors to ICIs therapy.21 The Cancer Immunome Atlas (TCIA) (http://tcia.at/home/) offers a download of the IPS values for NSCLC patients. In addition, based on 18 T cell inflammatory genes screened in previous studies, the T cell inflamed score was calculated by taking a weighted sum of the internal standardized values of the expression of these genes.22 We assessed the role of GBP5 in predicting response to anti-tumor immunotherapy by analyzing the correlation of GBP5 with the elements described above.
Clinical Samples
The NSCLC tissue and para-tumor tissue microarray (TMA) (Cat. HLuA150CS02, Cat. HLugS120CS01) were purchased from Outdo BioTech (Shanghai, China). A total of 118 NSCLC tissue and para-tumor samples were included in this study. The baseline characteristics could be found in Table S2. Detailed Clinicopathological features were obtained from Outdo BioTech.
Immunohistochemistry Staining and Semi-Quantitative Scoring
IHC staining included experimental procedures, including tissue dewaxing, antigen repair, inactivated enzyme, blocking, primary and secondary antibody incubation, color development and counterstaining, dehydration, plate sealing, and scanning. We stained the tissue microarrays used in this study by the IHC process described above. Sections were retrieved via EDTA. The primary antibodies used were anti-GBP5 (1:1000 dilution, Cat. 13220-1-AP, ProteinTech) and anti-PD-L1 (Ready-to-use, Cat.GT2280, GeneTech). Staining was observed with DAB and hematoxylin counterstain, and stained sections were acquired with the Aperio Digital Pathology Slide Scanners. Stained sections were analyzed semi-quantitatively by two pathologists, respectively, using immunoreactivity score (IRS)23 evaluation based on GBP5 and PD-L1 staining results. The percentage of positively stained cells was scored as 0–4: 0 (<5%), 1 (6–25%), 2 (26–50%), 3 (51–75%), and 4 (>75%). The staining intensity was scored as 0–3: 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). The IRS equals the percentages of positive cells multiplied by staining intensity.
Statistical Analysis
Statistical analysis and plotting with SPSS 27.0, R language 4.2.1, and GraphPad Prism 9.0 were performed. Statistical differences between the two groups were analyzed with parametric Student’s t-tests or non-parametric Mann–Whitney tests, and categorical variables were assessed with chi-square tests. Survival analysis was performed by Log rank test. The Pearson test was used to evaluate the correlation between two variables. All statistical tests were two-sided, and P value < 0.05 was considered statistically significant.
Results
Expression and Prognostic Value of GBP5 in NSCLC
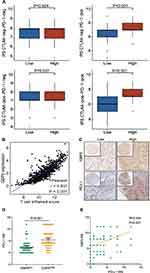
We used TCGA data to examine the mRNA levels of GBP5 in NSCLC tumor and para-cancer tissues. The findings demonstrated that GBP5 was upregulated in the tumor tissues (P=0.003) (Figure 1A). Meanwhile, analysis using the CTPAC dataset revealed that GBP5 protein was differentially expressed in the normal and primary tumor of LUAD (P=0.002) and was highly expressed in the primary tumor (Figure 1B). We carried out IHC staining and analysis of NSCLC TMA cohort to confirm the findings. According to the results, NSCLC tumor tissues had significantly higher levels of GBP5 expression than para-cancer tissues (P<0.001) (Figure 1C and D). Next, we estimated the predictive value of GBP5 on the prognosis of NSCLC with the help of the Kaplan–Meier plotter online tool. We discovered that patients with upregulated GBP5 expression had better overall survival (OS) (P=0.013, HR=0.81) (Figure 1E) and post-progression survival (PPS) (P=0.016, HR=0.59) (Figure 1F) than those with low expression, suggesting a better prognosis for NSCLC patients. These data suggested GBP5 as a potential prognosis biomarker for NSCLC.
GBP5 is Associated with Tumor Immune Microenvironment in NSCLC
Given the potential predictive value of GBP5 in NSCLC for prognosis, we have used the TCGA database to investigate whether the prognostic role of GBP5 in NSCLC is related to tumor immune-related properties. First, we used the TCGA database to discover that GBP5 positively correlated with most immunomodulators. There was a noticeable upregulation of many chemokines, receptors, MHC molecules, immunostimulators, and immunoinhibitors in the high-GBP5 group (Figure 2A). Chemokines and receptors in the tumor immune microenvironment can recruit effector TIICs, so it is hypothesized that GBP5 expression showed a positive correlation with increased TIICs in the tumor microenvironment (TME). As follows, we evaluated using two independent algorithms, TIMER and EPIC algorithms. We found that GBP5 was positively associated with TIICs abundance in NSCLC (Figure 2C). Also, GBP5 was found to be negatively associated with tumor purity (Figure 2B). Furthermore, GBP5 was found to be favorably associated with the expression level of immune checkpoint molecules (Figure 2D). All these results showed GBP5 was significantly related to anti-tumor immunity.
GBP5 Predicts the Response to Immunotherapy in NSCLC
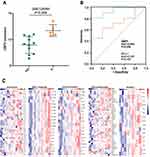
Based on the findings of the currently published articles and the above results, GBP5 protein expression was significantly correlated with immunity. We investigated GBP5 predictive value for NSCLC patients receiving immunotherapy in further detail. IPS can be used to predict how tumor patients will respond to immunotherapy. The median of GBP5 protein expression was used to classify them into high and low-expression groups. Our analysis showed that patients in the GBP5 high expression group had markedly higher IPS (Figure 3A). T cell inflamed score presents a profile composed of IFN-γ-related mRNAs that predict anti-PD-1 treatment efficacy. Our results show that GBP5 had a positive correlation with the T cell inflamed score (Figure 3B).
An NSCLC TMA cohort was included in this analysis to confirm the above results further. This cohort comprises 118 TMA samples with clinicopathological features representative of NSCLC that were used for analysis. Table 1 shows no statistical significance between samples with distinct GBP5 expression levels and age, Pathological type, TNM stage, T stage, N stage, M stage, and Differentiation, except for gender. TMA IHC results showed that GBP5 protein expression was notably upregulated in NSCLC tissues (Figure 1C and D). Furthermore, the NSCLC cohort was divided into low- and high-expression groups according to the median level of GBP5 IRS score (IRS<7 vs IRS≥8). We found that PD-L1 expression was higher in the GBP5 high expression group (P<0.001) (Figure 3C and D). Moreover, GBP5 was a positive association with PD-L1 expression in the NSCLC cohort (R=0.454, P<0.001) (Figure 3E). Combining the above findings, GBP5 can be used in combination with other immune microenvironment molecules for the immunophenotyping of NSCLC. The GBP5 high expression can identify inflamed TME and immuno-hot tumors.
 |
Table 1 The Relationship Between GBP5 Expression in NSCLC Patients |
We obtained the GSE126044 dataset from the GEO database to analyze the predictive value of GBP5 on the efficacy of immunotherapy. The expression of GBP5 was enhanced with the responder compared to the non-responder (P=0.004) (Figure 4A). Using the GSE126044 cohort, we analyzed the prediction of both GBP5 and PD-L1 on immunotherapeutic outcomes. It was shown that GBP5 had a higher predictive effect (AUC=0.945) than PD-L1 (AUC=0.727) (Figure 4B). In addition, we analyzed the correlations between GBP5 and the respective immunomodulators, which are the main components of TIME. The results demonstrated a positive correlation between GBP5 and most immunomodulators (Figure 4C). Combined with these results, GBP5 may be a potential biomarker for predicting the efficacy of immunotherapy in NSCLC.
GBP5 Correlates with Tumor Immune Microenvironment Across Cancer Types
Based on the TCGA dataset of NSCLC and GSE126044, the results of our analysis suggest that most immunomodulators in NSCLC are positively correlated with the expression of GBP5. To investigate the immune relevance of GBP5 in other tumors, we used the Sangerbox version 3.0 website single gene pan-cancer tools to examine the correlation of GBP5 with factors related to the TIME, such as chemokines, receptors, MHC, immunoinhibitors, and immunostimulators. The expression of GBP5 showed a positive correlation with the levels of these immunomodulatory molecules in various malignancies. (Figure 5A). Then, for many cancer types, GBP5 was linked negatively with tumor purity (Figure 5B) and positively with TIIC levels (Figure 5C). Additionally, the relationship between TMB (Figure 5D) and neoantigen burden (Figure S1) and GBP5 varied depending on the kind of malignancy. In conclusion, except for a few tumor types, GBP5 positively correlates with the tumor immune microenvironment and identifies the immunological phenotype of tumor tissues.
Discussion
NSCLC is one of the malignancies with the most significant morbidity and mortality rates. About 70% of NSCLC is diagnosed at an advanced stage, with a 5-year survival rate of about 23%.24 In the era of precision therapy, the emergence of targeted drugs targeting sensitive driver mutations and immunotherapy drugs represented by immune checkpoint inhibitors has opened a new era for the treatment of NSCLC and significantly prolonged the survival of patients. For NSCLC patients without driver mutation, chooses single-agent immunotherapy, immune combination chemotherapy, and dual immunotherapy according to immune checkpoint expression and organism condition. Currently, several immunotherapeutic agents are expected to be more widely used in the clinic, among which anti-CTLA-4 and anti-PD-1, and anti-PD-L1 therapy are the most prominent ones. Ipilimumab, which targets CTLA-4, has shown good efficacy in combination with radiotherapy or chemotherapy regimens.25,26 Pembrolizumab, which targets PD-1, has shown significant improvement in the patient prognosis when used as monotherapy.27 Sugemalimab, which targets PD-L1, has shown good efficacy in consolidation therapy for patients with advanced NSCLC.28 However, there are still some patients who do not respond to immunotherapy. With the advancement of research, it is gradually recognized that the heterogeneity and dynamic changes of TIME have an essential impact on immunotherapy response.
The TME is a dynamic system composed mainly of immune cells, stromal cells, tumor cells, and complex cytokines and chemokines. Immune and stromal cells are the two major non-tumor components of the TME and are thought to play an essential role in diagnosing and prognosis of tumors.29 Based on the distribution of immune cells in the TME, tumor tissues can be classified into three immune phenotypes: inflamed, excluded, and desert phenotypes.30 Due to the predominance of immuno-suppressive TME and lack of immune cell infiltration in the microenvironment, the latter two phenotypes are called “cold” tumors, which are mostly ineffective for immunotherapy. Immune-inflamed tumors, also known as “hot” tumors, are featured by high T-cell infiltration, enhanced IFN-γ signaling, PD-L1 expression, and high TMB, and tumors with inflammatory phenotypic features are more sensitive to immunotherapy.31 Therefore, exploring immune-related factors in the TME and identifying potential biomarkers associated with TIME features is vital for screening beneficiaries of immunotherapy in clinical practice.
PD-L1 is a prevalent clinical predictor of treatment for NSCLC ICIs due to its strong efficacy as an immunotherapy biomarker.32 However, clinical data results show limitations if PD-L1 alone is used as an indicator to screen patients suitable for ICIs treatment. Previous studies suggested that IFN-γ significantly enhances PD-L1 expression, while PD-L1 is upregulated in immune “hot” tumors and significantly correlates with the characteristics of TIME.33 GBP5, a downstream gene of IFN-γ induced expression, correlates well with PD-L1 expression.
GBP5 belongs to the family of IFN-γ-inducible large GTPases and is responsible for many cellular functions, and it has been found to promote the activation of NLRP3-dependent inflammatory responses.14 GBP5 activates the Src/ERK1/2 MAPK pathway to induce matrix metalloproteinase 3 (MMP3) expression, which plays a crucial role in the malignant biological behavior of GBM tumor cells.12 The expression of many genes in colon cancers is associated with anti-tumor immunity, including GBP1, GBP4, GBP5, NKG7, APOL3, IDO1, CCL5, and CXCL9.11 Activation of the TNF-α/NF-κB signaling axis in lung cancer promotes EMT progression and PD-L1 expression, and upregulation of GBP5 in triple-negative breast cancers (TNBCs) may be associated with activation of the TNF-α/NF-κB signaling pathway. Therefore, we hypothesized that the upregulation of GBP5 may predict the effect of immune checkpoint blockade in treating TNBC.34,35 Previous studies have shown GBP5 is associated with a good prognosis for basal-like breast tumors with high programmed cell death protein 1 (PD-1) and PD-L1 expression and is associated with immune infiltration.36 In addition, the IFN-γ score consisting of seven IFN-γ inducible genes in glioma, namely GBP5, ICAM1, CAMK2D, IRF1, SOCS3, CD44, and CCL2, could be used as an adjuvant prognostic indicator for screening patients suitable for ICIs.33 However, further studies in NSCLC are still lacking.
Based on RNA-Seq data onto online datasets and validation of the NSCLC TMA of IHC, GBP5 was highly expressed in NSCLC cells compared with para-cancer tissue, and high GBP5 expression had better OS and PPS than those with low expression. Our results show that GBP5 positively correlates with most immune-related genes, TIIC levers, and PD-L1 expression. This suggests the immune system is more active in tumors with upregulated GBP5 expression, which may be considered immunologically “hot” tumors. We then further analyzed the prediction of GBP5 for immunotherapy efficacy in NSCLC undergoing immunotherapy and showed that GBP5 was positively correlated with most immunomodulators. Its predictive value was significantly higher than that of PD-L1.
In summary, this study revealed that GBP5 immunological significance and predictive value for immunotherapy response in NSCLC datasets revealed GBP5 to be a potential biomarker that makes it easier to predict the effectiveness of immunotherapy in NSCLC. However, this study lacked clinical tissue microarray of NSCLC patients before and after immunotherapy. Only a series of analyses of GBP5 expression and immune correlation characteristics and prognostic value were performed using public data from the internet. Therefore, subsequent clinical tissue microarrays need to be collected to verify the potential value of GBP5 for NSCLC patients receiving immunotherapy. Meanwhile, the number of cases in the immunotherapy cohort included in the current study is limited. Further validation in patients receiving immunotherapy with ICIs on a large scale is needed.
Abbreviations
ICI, immune checkpoint inhibitor; NSCLC, non-small cell lung cancer; GBP5, guanylate binding protein 5; TCGA, The Cancer Genome Atlas; CPTAC, Clinical Proteomic Tumor Analysis Consortium; PD-L1, Programmed cell death 1 ligand 1; TMB, tumor mutational burden; MSI, microsatellite instability; IFN-γ, Interferon-gamma; GBM, glioblastoma; FPKM, fragments per kilobase per million; LUAD, lung adenocarcinoma; IHC, immunohistochemistry; GEO, Gene Expression Omnibus; TIME, tumor immune microenvironment; TIIC, tumor-infiltrating immune cell; MHC, major histocompatibility complex; IPS, immunophenoscore; TCIA, The Cancer Immunome Atlas; TMA, tissue microarray; IRS, immunoreactivity score; OS, overall survival; PPS, post-progression survival; TME, tumor microenvironment; MMP3, matrix metalloproteinase 3; TNBC, triple-negative breast cancer; PD-1, programmed cell death protein 1.
Data Sharing Statement
All data supporting the results of this study are shown in this published article and Supplementary Documents. In addition, original omics data for bioinformatics analysis could be obtained from corresponding platforms.
Ethics Approval
Ethical approval (No. YB-M-05-02) for the use of TMAs was granted by the Clinical Research Ethics Committee in Outdo Biotech (Shanghai, China). Our study complies with the Declaration of Helsinki.
Acknowledgments
We thank those authors who released and shared their datasets on the TCGA, CPTAC, and GEO databases.
Author Contributions
Chaoying Liu and Junying Xu designed the research and the experiments. Huiyu Wang, Yuting Li, and Jie Mei conducted data analysis and data interpretation; Honghong Fan and Yuxin Shi wrote the manuscript and completed statistical analysis. All listed authors must have made a significant scientific contribution to the research in the manuscript approved its claims and agreed to be an author. All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Wuxi Science and Technology Bureau Foundation (No. N20202018). Beijing Hengji Health Management and Development Foundation (HJ-HX-ZLXD-202209-002).
Disclosure
The authors declare that they have no competing interests.
References
1. Hamid O, Robert C, Daud A, et al. Five-year survival outcomes for patients with advanced melanoma treated with pembrolizumab in KEYNOTE-001. Ann Oncol. 2019;30(4):582–588. doi:10.1093/annonc/mdz011
2. Das S, Johnson DB. Immune-related adverse events and anti-tumor efficacy of immune checkpoint inhibitors. J Immunother Cancer. 2019;7(1):306. doi:10.1186/s40425-019-0805-8
3. Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16(5):275–287. doi:10.1038/nrc.2016.36
4. Marabelle A, Fakih M, Lopez J, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, Phase 2 KEYNOTE-158 study. Lancet Oncol. 2020;21(10):1353–1365. doi:10.1016/S1470-2045(20)30445-9
5. Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. doi:10.1186/s13073-017-0424-2
6. Zhu Z, Chen P, Yan Z. DNA damage response signaling as a predictive biomarker and synergistic therapeutic target for anti-PD-1/PD-L1 immunotherapy in non-small cell lung cancer. Thorac Cancer. 2018;9(8):901–903. doi:10.1111/1759-7714.12785
7. Vestal DJ, Jeyaratnam JA. The guanylate-binding proteins: emerging insights into the biochemical properties and functions of this family of large interferon-induced guanosine triphosphatase. J Interferon Cytokine Res. 2011;31(1):89–97. doi:10.1089/jir.2010.0102
8. Braun E, Hotter D, Koepke L, et al. Guanylate-binding proteins 2 and 5 exert broad antiviral activity by inhibiting furin-mediated processing of viral envelope proteins. Cell Rep. 2019;27(7):2092–104.e10. doi:10.1016/j.celrep.2019.04.063
9. Li P, Jiang W, Yu Q, et al. Ubiquitination and degradation of GBPs by a Shigella effector to suppress host defence. Nature. 2017;551(7680):378–383. doi:10.1038/nature24467
10. Patil PA, Blakely AM, Lombardo KA, et al. Expression of PD-L1, indoleamine 2,3-dioxygenase and the immune microenvironment in gastric adenocarcinoma. Histopathology. 2018;73(1):124–136. doi:10.1111/his.13504
11. Elsayed I, Elsayed N, Feng Q, Sheahan K, Moran B, Wang X. Multi-OMICs data analysis identifies molecular features correlating with tumor immunity in colon cancer. Cancer Biomark. 2022;33(2):261–271. doi:10.3233/CBM-210222
12. Yu X, Jin J, Zheng Y, et al. GBP5 drives malignancy of glioblastoma via the Src/ERK1/2/MMP3 pathway. Cell Death Dis. 2021;12(2):203. doi:10.1038/s41419-021-03492-3
13. Shaverdian N, Lisberg AE, Bornazyan K, et al. Previous radiotherapy and the clinical activity and toxicity of pembrolizumab in the treatment of non-small-cell lung cancer: a secondary analysis of the KEYNOTE-001 Phase 1 trial. Lancet Oncol. 2017;18(7):895–903. doi:10.1016/S1470-2045(17)30380-7
14. Shenoy AR, Wellington DA, Kumar P, et al. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 2012;336(6080):481–485. doi:10.1126/science.1217141
15. Li G, Kryczek I, Nam J, et al. LIMIT is an immunogenic lncRNA in cancer immunity and immunotherapy. Nat Cell Biol. 2021;23(5):526–537. doi:10.1038/s41556-021-00672-3
16. Mei J, Cai Y, Xu R. Protocol to identify novel immunotherapy biomarkers based on transcriptomic data in human cancers.STAR Protoc. 2023;4(2):102258. doi: 10.1016/j.xpro.2023.102258
17. Mei J, Fu Z, Cai Y. SECTM1 is upregulated in immuno-hot tumors and predicts immunotherapeutic efficacy in multiple cancers. iScience. 2023;26(2):106027. doi: 10.1016/j.isci.2023.106027
18. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e10. doi:10.1158/0008-5472.CAN-17-0307
19. Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218. doi:10.1186/s13059-016-1070-5
20. Shen W, Song Z, Zhong X, et al. Sangerbox: a comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta. 2022;1(3):e36. doi:10.1002/imt2.36
21. Charoentong P, Finotello F, Angelova M, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. doi:10.1016/j.celrep.2016.12.019
22. Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. doi:10.1172/JCI91190
23. Cai Y, Ji W, Sun C, et al. Interferon-induced transmembrane protein 3 shapes an inflamed tumor microenvironment and identifies immuno-hot tumors. Front Immunol. 2021;12:704965. doi:10.3389/fimmu.2021.704965
24. Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69(5):363–385. doi:10.3322/caac.21565
25. Wilkins A, McDonald F, Harrington K, Melcher A. Radiotherapy enhances responses of lung cancer to CTLA-4 blockade. J Immunother Cancer. 2019;7(1):64. doi:10.1186/s40425-019-0542-z
26. Lynch TJ, Bondarenko I, Luft A, et al. Ipilimumab in combination with paclitaxel and carboplatin as first-line treatment in stage IIIB/IV non-small-cell lung cancer: results from a randomized, double-blind, multicenter Phase II study. J Clin Oncol. 2012;30(17):2046–2054. doi:10.1200/JCO.2011.38.4032
27. Leighl NB, Hellmann MD, Hui R, et al. Pembrolizumab in patients with advanced non-small-cell lung cancer (KEYNOTE-001): 3-year results from an open-label, phase 1 study. Lancet Respir Med. 2019;7(4):347–357. doi:10.1016/S2213-2600(18)30500-9
28. Zhou Q, Chen M, Jiang O, et al. Sugemalimab versus placebo after concurrent or sequential chemoradiotherapy in patients with locally advanced, unresectable, stage III non-small-cell lung cancer in China (GEMSTONE-301): interim results of a randomised, double-blind, multicentre, Phase 3 trial. Lancet Oncol. 2022;23(2):209–219. doi:10.1016/S1470-2045(21)00630-6
29. Jia D, Li S, Li D, Xue H, Yang D, Liu Y. Mining TCGA database for genes of prognostic value in glioblastoma microenvironment. Aging. 2018;10(4):592–605. doi:10.18632/aging.101415
30. Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–330. doi:10.1038/nature21349
31. Liu YT, Sun ZJ. Turning cold tumors into hot tumors by improving T-cell infiltration. Theranostics. 2021;11(11):5365–5386. doi:10.7150/thno.58390
32. Hurkmans DP, Kuipers ME, Smit J, et al. Tumor mutational load, CD8(+) T cells, expression of PD-L1 and HLA class I to guide immunotherapy decisions in NSCLC patients. Cancer Immunol Immunother. 2020;69(5):771–777. doi:10.1007/s00262-020-02506-x
33. Qian J, Wang C, Wang B, et al. The IFN-γ/PD-L1 axis between T cells and tumor microenvironment: hints for glioma anti-PD-1/PD-L1 therapy. J Neuroinflammation. 2018;15(1):290. doi:10.1186/s12974-018-1330-2
34. Asgarova A, Asgarov K, Godet Y, et al. PD-L1 expression is regulated by both DNA methylation and NF-kB during EMT signaling in non-small cell lung carcinoma. Oncoimmunology. 2018;7(5):e1423170. doi:10.1080/2162402X.2017.1423170
35. Cheng SW, Chen PC, Lin MH, Ger TR, Chiu HW, Lin YF. GBP5 repression suppresses the metastatic potential and PD-L1 expression in triple-negative breast cancer. Biomedicines. 2021;9(4):371. doi:10.3390/biomedicines9040371
36. Cimas FJ, Manzano A, Baliu-Piqué M, et al. Genomic mapping identifies mutations in RYR2 and AHNAK as associated with favorable outcome in basal-like breast tumors expressing PD1/PD-L1. Cancers. 2020;12(8):2243. doi:10.3390/cancers12082243
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.