Back to Journals » OncoTargets and Therapy » Volume 13
Functional Implication of Exosomal miR-217 and miR-23b-3p in the Progression of Prostate Cancer
Authors Zhou C, Chen Y, He X, Zheng Z , Xue D
Received 31 July 2020
Accepted for publication 28 October 2020
Published 12 November 2020 Volume 2020:13 Pages 11595—11606
DOI https://doi.org/10.2147/OTT.S272869
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Carlos E Vigil
Cuixing Zhou,1 Yimeng Chen,1 Xiaozhou He,1 Zhuojun Zheng,2 Dong Xue1
1Department of Urology, The Third Affiliated Hospital of Soochow University, Changzhou, People’s Republic of China; 2Department of Hematology, The Third Affiliated Hospital of Soochow University, Changzhou, People’s Republic of China
Correspondence: Dong Xue
Department of Urology, The Third Affiliated Hospital of Soochow University, Changzhou, Jiangsu Province, People’s Republic of China
Tel +86-519-6887 1257
Email [email protected]
Zhuojun Zheng
Department of Hematology, The Third Affiliated Hospital of Soochow University, Changzhou, Jiangsu Province, People’s Republic of China
Tel +86-519-6887 1092
Email [email protected]
Objective: The microRNA expression profile of plasma exosomes in prostate cancer (PCa) is of critical importance in the disease exploration. This study aimed to explore the clinical application of exosomal miRNAs as biomarkers for PCa.
Methods: Exosome-like vesicles of PCa patients and healthy controls were purified by differential centrifugation. The purified vesicles within the ranges of 50 and 100 nm were classified as exosomes according to the results of transmission electron microscopy and Western blot. Both, in vitro and in vivo, validations were performed by small RNA sequencing, CCK8, RT-qPCR, flow cytometry, Western blot, transwell and immunofluorescent staining assays.
Results: High-throughput sequencing identified that 94 miRNAs were differentially expressed in PCa patients in comparison with healthy controls (P< 0.01; fold change ≥ 2). Among them, 64 miRNAs were upregulated, and 30 miRNAs were downregulated. In comparison to the healthy controls, the expression levels of miR-217 were significantly upregulated, while miR-23b-3p were significantly downregulated in the exosomes and serum collected from PCa patients. Both, in vitro and in vivo, studies revealed that exosomes secreted by PCa cells with up-regulated miR-217 levels promoted cell proliferation and invasion; meanwhile, the exosomes with up-regulated miR-23b-3p levels inhibited cell proliferation and invasion. The epithelial–mesenchymal transition process may have been involved in the above-mentioned regulation.
Conclusion: This study identified the dysregulated expression of exosomal miRNAs in PCa patients, including miR-217 and miR-23b-3p, by validating their function on proliferation and invasion in PCa cells. This regulation may have been affected by the epithelial–mesenchymal transition process, suggesting that they can be used as potential targets in the diagnosis and treatment of PCa.
Keywords: exosome, miR-217, miR-23b-3p, prostate cancer
Introduction
Prostate cancer (PCa) is one of the most common malignant tumors in the male genitourinary system. In Europe and the United States, PCa has the highest incidence rate and the third highest mortality rate among male malignant tumors. In recent years, the incidence of PCa has increased gradually.1 However, due to the lack of effective procedures to diagnose early, 80% of the patients in China were diagnosed with bone metastasis upon their first diagnosis.2 The situation in England,3 Canada,4 and the US5 is much better, since screening is more widespread. Still, PCa is a very dangerous type of malignancy. Clinically, common treatment strategies for PCa include surgery, radiation therapy, and endocrine therapy. Endocrine therapy, such as surgical or medical castration and androgen receptor antagonist, or other antiandrogen drug treatments, has been the only treatment for advanced metastatic PCa.6 However, according to previous studies, almost every endocrine therapy-sensitive patient eventually develops glucocorticoid resistance, leading to resistance to the endocrine treatment, advancement in the development of cancer, which causes irreversible clinical progress, and subsequently death of patients.7 With the development of molecular biology and related fields, gene therapy nowadays is becoming an important approach in the early diagnosis and treatment of PCa, meanwhile biomarkers followed by effective targeted therapies are also promising. Therefore, research on the discovery and identification of new biological markers is urgently needed.
Exosomes were first discovered in the late 80s by Johnstone et al during their study of vesicle formation in mature reticulocytes.8 Several studies on exosomes have been published thereafter. The exosome is described as a spherical vesicle carrier with a diameter between 30 and 100 nm, and a density between 1.10 and 1.18 g/mL, which is seen as a flat, spherical, or cup-shaped corpuscle under an electron microscope.9 During the formation of exosomes, polyvesicular corpuscles are formed after a series of complex mechanisms, following membrane invagination. Subsequently, the polyvesicle’s intracellular membrane fuses with the cell membrane to release exosomes. Exosomes have a double-capsule-membrane structure, and are rich in lipids, proteins and nucleic acids. As of 2020, according to the ExoCarta database (http://www.exocarta.org/), a total number of 9769 proteins, 3408 mRNA, 2838 miRNA, and 116 lipids have been discovered in exosomes. It was proposed that exosomes, secreted by certain tumor cells, potentially carry cell adhesion molecules, metalloproteinases, and tissue-specific proteins, suggesting a close relationship with the occurrence and development of tumors.10,11 In addition, exosomes are abundant in nucleic acids, among which miRNAs and mRNAs were the first to be discovered, while tRNA, lncRNA, viral RNA, and other types have been discovered in further research.12
Exosomes secreted by tumor cells are referred to as texosomes (TEX), which is an important component in the tumor microenvironment. TEX potentially participate, in the anti-tumorigenicity of proto-oncogenes and the formation of the internal environment during the anti-tumor process, through the intercellular transmission responses of the host, thus affecting the processes of tumor occurrence, development, invasion, metastasis, angiogenesis, immune responses and tumor escape.13 In our study, we performed miRNA profiling of plasma exosomes from patients with PCa and healthy controls through high-throughput sequencing. We further, in vitro and in vivo, selected, analyzed, and validated specific miRNAs in PCa cells.
Materials and Methods
Sample Acquisition
Written informed consent was obtained from all participants prior to the start of this study.10 patients with PCa were recruited for plasma sample collection at the 3rd Affiliated Hospital of Soochow University during the period Jan 2016 to Jan 2018. The patients were between 54 and 79 years old with an average age of 68.27 years-, and standard deviation (SD) of 4.59 years. The characteristics of included patients are summarized in Table 1. All patients underwent biopsy and the pathological diagnosis confirmed positive PCa. None of the patients had ever received any radical or curative treatments and were all diagnosed with stage III PCa. Patients with any predisposing systemic disorders were excluded. Another 10 healthy participants were recruited from the physical examination center as the control group within the above-mentioned period of time. The control group had a comparable age range of 47 to 78 years old, with an average age of 63.83 years- and a SD of 6.53 years. Ten-milliliter EDTA tubes were used for the collection of blood, by venipuncture, from the patients at fasting state. The blood samples underwent centrifugation at 2000 g for 10 min at 4°C. The extracted samples were stored as 750 μL aliquots at −80°C. The experiment was approved by the Institutional Human Experiment and Ethics Committee of the 3rd Affiliated Hospital of Soochow University. The study was conducted according to the Declaration of Helsinki.
 |
Table 1 Baseline of Patients Included in This Study |
Cell Culture
PCa cells (PC-3 and DU145) derived from humans were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum and 5% penicillin-streptomycin, and incubated at 37°C with 5% CO.2
Exosome Preparation and Size Determination
At 4°C, the plasma samples were centrifuged at 1600 g for 20 min and at 10,000 g for 30 min. The extracted supernatant was processed with 0.22 μm disposable filter units. The resulting filtration was centrifuged at 100,000 g and 4°C for 2 h. Pellets containing exosomes were washed with 10 mL PBS, resuspended in 0.1 mL PBS and kept at −80°C for further experiments. The diameter of the exosomes was measured with Zetasizer Nano in compliance with the manufacturer’s instructions. An EM-2010 transmission electron microscope (TEM) at x250,000 magnification was used for further examination of the prepared exosome samples.
Western Blot
Protein samples were prepared in RIPA lysis buffer (Abcam, ab156034 with protease inhibitor cocktail). The protein concentration of the samples was tested with a BCA protein assay kit (Abcam, ab102536). Aliquots of 20 μg protein was fractionated with 10% SDS-PAGE gel and transferred to nitrocellulose membranes. The membranes were blocked in 5% skimmed milk, dissolved in PBST (0.5% Tween-20) buffer for 2 h at room temperature, and incubated with the following primary antibodies: rabbit anti-TSG101 (Abcam, ab125011, 1/1000), mouse anti-HSP70 (Abcam, ab181606, 1/1000), rabbit anti-E-cadherin(ab15148) (1/500), or rabbit anti-Vimentin (ab92547) (1/1000) at 4°C overnight. Secondary antibodies (1/1000) were subsequently incubated with the membranes at room temperature for 1 h. Positive staining was visualized on X-ray films with Pierce enhanced chemiluminescent visualization reagents.
Exosomal RNA’s Isolation and Small RNA Sequencing
To obtain exosomal total RNA from the plasma samples, RNeasy Mini Spin kit (QIAGEN, Cat No./ID: 74104) was applied as per manufacturer’s instructions. The concentration and purity of the RNA extractions were detected with the Agilent 2100 bioanalyzer system. Small RNA sequencing was conducted with the BGISEQ-500 sequencer. Significantly interfered results were ignored, and distinctive adaptor sequences or RNA small sequences, which had perfect matches in the human genome database (http://ncbi.nlm.nih.gov/genomes/Homo_sapiens), were deemed valid for the subsequent analytical procedures. Based on the seed sequences, target mRNAs were identified with TargetScan (version 7.2; http://www.targetscan.org/vert_72/). The expression levels of miRNA were analyzed according to the frequency of detection with the DEGseq method.14
Target Gene and Pathway Enrichment Analysis
Target genes for the miRNAs of interest were researched within three databases: TargetScan,15 miRanda16 and RNAhybrid.17 The pathway analysis of the target genes included Gene Ontology (GO) functional annotation18 and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis,19 and was performed with DIANA TOOLS (http://snf-515788.vm.okeanos.grnet.gr/).
Plasma RNA Extraction
The total RNA of the plasma samples was extracted with miRNeasy Serum/Plasma kit in accordance with the manufacturer’s instructions. After extracting, it was suspended in 14 μL nuclease-free water. The concentration of the RNA samples was measured with a NanoDrop ND-2000 spectrophotometer.
Quantitative Polymerase Chain Reaction (qPCR)
cDNA was reverse transcribed from total RNA with the Qiagen miScript II RT kit and qPCR was conducted with the Qiagen miScript SYBR Green PCR kit, both in accordance with the manufacturer’s instructions. The miScript primers were used as shown in Table 2. The Cq values were normalized to synthetic spike as described previously.20 Relative fold-changes were estimated with the 2-ΔΔCq method.21
 |
Table 2 miScript Primers Used in This Study |
Cell Transfection
50 nm miRNA-217 mimic and miRNA-23b-3p mimic were transfected into the PC-3 and DU145 cells using Lipofectamine™ 2000 (Invitrogen). The exosomes were isolated from transfected cells for further validating assays. miRNA-217 mimic and miRNA-23b-3p mimic were obtained from GenePharma (GenePharma Corporation, Shanghai, China).
Cell Proliferation Assay
PCa cells that were stimulated with negative control (NC), tumor-secreted exosomes, derived from normal PCa cells (PC-3 and DU145), and tumor-secreted exosomes derived from miR-217 or miR-23b-3p up-regulated PC-3 and DU145 cells, were seeded in 96-well culture plates. Next, the respective levels of cell proliferation were measured with CCK-8 (Dojindo, Japan) following the manufacturer’s instructions, at the timepoints 24, 48, 72, and 96 h onwards.
Flow Cytometry
Apoptosis analysis was performed with the Annexin V-FITC kit as previously described. In brief, PCa cells stimulated with NC, tumor-secreted exosomes derived from PC-3 and DU145 cells, and tumor-secreted exosomes derived from miR-217 or miR-23b-3p up-regulated PC-3 and DU145 cells, were used for flow cytometry analysis. The derived data were processed with CellQuest software (BD Biosciences). Cell cycle distribution was analyzed by FACS Calibur flow cytometer (BD).
Transwell Assay
The evaluation of cellular mobility was performed by transwell assays. In brief, the above-mentioned treated PCa cells were incubated in the upper chamber of the transwell chambers (Corning costar, 8-μm pore size; Cambridge, USA), while 20% serum, serving as chemoattractant, was added into the lower chamber. After incubation for 48 h, the filter was fixed with methanol and 0.1% crystal violet, and stained. The cells were measured with a microscope on the bottom side of the filter. Each trial was conducted in triplicates.
Immunofluorescent Staining (IF)
Briefly, the PCa cells were orderly incubated with rat monoclonal anti-E-cadherin (Abcam, ab11512, 10 µg/mL), mouse monoclonal anti-Vimentin (Abcam, ab8978, 1 µg/mL), Alexa®488-conjugated goat anti-rabbit secondary antibody (Thermo Fisher, CA, USA), and the nuclear stain Hoechst 34,580 (Molecular Probes, Thermo Fisher, CA, USA, 5 μg/mL). Images were obtained by using the Zeiss confocal microscope.
In vivo Assay
The animal experiments were approved by the Institutional Animal Care and Use Committee of the 3rd Affiliated Hospital of Soochow University. NIH principles of laboratory animal care were strictly adhered to guarantee welfare of the animals. PC-3 (1 × 106) and DU145 (1 × 106) cells with different exosome-treatment were subcutaneously inoculated into the left upper, right upper, and right lower limbs of 4-week-old BALB/C nude mice. After 4 weeks, the mice were sacrificed, and the xenografts were separated, weighed and measured. The volume was calculated according to the following formula: volume (cm3) = (width2 ×length)/2.
Immunohistochemistry (IHC)
From formalin-fixed and paraffin-embedded blocks, 4-micron sections were cut and placed on charged slides. The sections were pre-treated using heat-mediated antigen retrieval with sodium citrate buffer (pH 6, epitope retrieval solution 1) for 20 min. Rabbit monoclonal Vimentin antibodies (Abcam, ab92547, 1/200 dilution) were incubated for 15 min and detected using a HRP-conjugated compact polymer system. DAB was used as the chromogen. Then, the sections were counterstained with haematoxylin and mounted with DPX. An IHC score was assigned, by two pathologists, according to the following criteria: 3+, intense, granular cytoplasmic staining; 2+, moderate, smooth cytoplasmic staining; 1+, faint cytoplasmic staining in ≥10% of tumor cells; and 0, no staining.
Statistical Analysis
GraphPad Prism 5 was used for all the statistical analyses. The comparison of two groups of data was conducted with the unpaired one-tailed t-test. P<0.05 was defined as statistically significant.
Results
Plasma Exosomes Isolated from PCa Patients
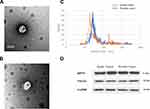
The age ranges of the two groups, 10 patients with PCa and 10 healthy controls, were comparable without a statistically significant inter-group difference (P>0.05). TEM images (Figure 1A and B) revealed that the plasmatic extracellular vehicles (EVs), in both groups, were spherical vesicles with diameters ranging between 50 and 100 nm, which were similar to that of the exosomes (Figure 1C). Moreover, two identified exosome markers, HSP70 and TSG101, were present in the EVs from both groups (Figure 1D).
Differentially Expressed miRNAs in Exosomes
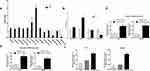
With the settings of 2-fold change and P<0.01 as cutoff value, the expression of 94 miRNAs of the exosomes collected from the PCa patients were significantly different compared to the healthy controls. Among them, 64 miRNAs had increased expression and 30 miRNAs had decreased expression in the exosomes. The 10 miRNAs with most upregulated expression include: hsa-miR-659-3p, hsa-miR-18a-5p, hsa-miR-127-5p, hsa-miR-941, hsa-miR-147a, hsa-miR-422a, hsa-miR-4704-5p, hsa-miR-4799-5p, hsa-miR-663a, and hsa-miR-551b-5p. In addition, the 10 miRNAs with most downregulated expression include: hsa-miR-29c-3p, hsa-miR-519a-3p, hsa-miR-636, hcmv-miR-UL70-5p, hsa-miR-4255, hsa-miR-4755-5p, hsa-miR-606, hsa-miR-5571-3p, hsa-miR-3064-5p, and hiv1-miR-TAR-3p. The heatmap and volcano plot with red and green spots to indicate the miRNAs with significantly different levels of expression, are shown in Figure 2A and B.
Enriched Biological Processes and Molecular Functions (KEGG) Analysis
KEGG pathway analysis demonstrated that most of the relevant genes were primarily involved as miRNAs in cancer and phagosomes in mucin-type O-Glycan biosynthesis, metabolism of xenobiotics by cytochrome P450 as well as the TGF-beta signaling pathway. GO analysis for the relevant genes of the miRNAs of interest demonstrated that, most of them related to the functions of positive regulation of cell migration, cellular nitrogen compound metabolic process, organelle, biosynthetic process, cell adhesion, small molecule metabolic process, catabolic process, collagen catabolic process and extracellular matrix organization. Above-mentioned results are shown in Figure 2C. Interestingly, the TGF-beta signaling pathway, positive regulation of cell migration and cell adhesion were found to be involved in the differentially expressed exosomal miRNAs, indicating that the regulation of epithelial-to-mesenchymal transition (EMT) potentially exists in the PCa serum exosome.
miRNA Expression Level
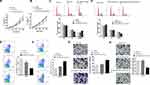
According to miRNA profiling, the relative expression levels of miR-147a, miR-659-3p, miR-520a-3p, let-7d-5p, miR-127-5p, miR-217, miR-422a, miR-519a-3p, miR-606, miR-148a-3p, miR-506-5p, and miR-23b-3p were investigated. Among them, miR-147a, miR-659-3p, miR-520a-3p, let-7d-5p, miR-127-5p, miR-217, and miR-422a were found to be upregulated and miR-519a-3p, miR-606, miR-148a-3p, miR-506-5p, and miR-23b-3p were downregulated in plasma exosomes of the PCa patients. Further confirmed by qPCR, miR-127-5p and miR-217 were upregulated, while miR-148a-3p and miR-23b-3p were significantly downregulated in the exosomes of PCa patients, in comparison to the healthy controls (Figure 3A). The level of miR-127-5p, miR-217, miR-148a-3p and miR-23b-3p were also measured in the serum of PCa patients. The results indicated that miR-217 was upregulated and miR-23b-3p downregulated (Figure 3B). In addition, the elevated expression of miR-217 and miR-23b-3p were confirmed, in PCa cells and exosomes derived from the corresponding PCa cells, when transfected with each miRNA mimic (Figure 3C and D). The normal PC-3 cells were treated with NC and exosomes were derived from normal PC-3 cells and miR-217-overexpressed PC-3 cells, while normal DU145 cells were treated with NC and exosomes were derived from normal DU145 cells and miR-23b-3p-overexpressed DU145 cells. The level of miR-217 and miR-23b-3p was examined in the aforementioned groups, respectively. As expected, miR-217 and miR-23b-3p were more expressed in miR-217-overexpressed PC-3 exosome treated PC-3 cells and miR-23b-3p-overexpressed DU145 exosome treated DU145 cells, respectively, compared to the control groups (Figure 3E).
Proliferation and Invasion of Exosomal miR-217 and miR-23b-3p Regulated PCa Cells
Gain-of-function assays were carried out to investigate the carcinogenic or carcinostatic role of miR-217 and miR-23b-3p derived from PCa-secreted plasma exosomes. The PC-3 and DU145 cells were treated as shown in Figure 3E. The cell proliferation in the miR-217-mimic PC-3 exosomes group was increased (Figure 4A), while it was decreased in the miR-23b-3p-mimic DU145 exosomes group (Figure 4B), compared with the corresponding control groups. Promoted G2/M cycle (Figure 4C) and inhibited cell apoptosis rates (Figure 4E), and inhibited G2/M cycle (Figure 4D) and enhanced cell apoptosis rates (Figure 4F), were demonstrated in the miR-217-mimic PC-3 exosomes group and miR-23b-3p-mimic DU145 exosomes group, respectively. Treatment with miR-217-mimic PC-3 exosomes enhanced the cell invasive capacity in the PC-3 cells (Figure 4G), while it diminished the cell invasive capacity in DU145 cells when treated with miR-23b-3p-mimic DU145 exosomes, which was proven by the transwell assay (Figure 4H). These differences were statistically significant (P<0.05).
Exosomal miR-217 and miR-23b-3p Regulate E-Cadherin, N-Cadherin and Vimentin Expression in PCa Cells
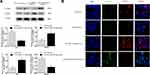
To investigate whether exosomal miR-217 and miR-23b-3p affect the EMT in PCa cells, PC-3 and DU145 cells were treated with NC and miR-217-mimic PC-3 exosomes or miR-23b-3p-mimic DU145 exosomes, respectively. Western blot assay was conducted for the expression of E-cadherin, and Vimentin. The results showed that the expression of E-cadherin was enhanced and the expression of Vimentin was restrained in the miR-23b-3p-mimic DU145 exosomes group, and vice versa in the miR-217-mimic PC-3 exosomes group (Figure 5A). These results were also confirmed by IF (Figure 5B), which preliminarily supported our hypothesis.
Validating Assays in vivo
The xenografts treated with miR-217-mimic exosomes were larger in size and heavier in weight compared to the ones of the Up-regulated-miR-23b-3pDU145 exosomes group (Figure 6A). The expression of miR-217, miR-23b-3p, and Vimentin, in removed tumor tissues, were analyzed by qPCR and IHC. As shown in Figure 6B, miR-217 and miR-23b-3p were more up-regulated in tumor tissue of the miRNA-mimic exosome treated group than other groups. Vimentin was more expressed in tissues of the miR-217-mimic PC-3 exosomes treated group and less expressed in tissues of the miR-23b-3p-mimic DU145 exosomes treated group compared to other groups (Figure 6C). These differences were statistically significant (P<0.05).
Discussion
Despite the important regulatory function of serum exosome miRNA in PCa, interest in this area is still limited. In our study, we compared exosome miRNA expression, in the serum of healthy subjects and PCa patients, to identify a characteristic pattern of miRNAs that could be used as a standard criterion for “liquid biopsy” in this specific patient population. In our analysis, a total of 64 up-regulated and 30 down-regulated miRNAs were discovered in the circulating exosomes, which could be related to tumor cell proliferation. Among them, miR-217 and miR-23b-3p were demonstrated, through a series of in vitro and in vivo assays, to regulate the proliferation and invasion ability of PCa cells via EMT. This novel finding contributes to the potential establishment of criteria for “liquid biopsy” in PCa. Neoplastic miRNAs of tumor cells could be transferred to other sites through exosomes, resulting in uncontrolled proliferation of tumor cells beyond the primary site (metastasis). On the other hand, tumor cells remain in the state of proliferation by eliminating the miRNAs of tumor suppressors. A previous study by Josson et al demonstrated that miR-409 in the PCa exosome was able to inhibit the process of enhancement in the formation of MET by the RAS suppressor protein 1 (RSU1) and matrix antigens.22 Other studies also found that miR-21 and miR-141 in the exosomes of PCa patients were able to regulate the activation of osteoclasts and osteoblasts in the tumor microenvironment, which may contribute to the promotion of bone metastasis.23,24 Using RNA expression microarray, Corcoran et al25 discovered four miRNAs, miR-598, miR-34a, miR-146a and miR-148a, that participated in the docetaxel resistance of PCa. Among which, miR-34a contributed to the docetaxel resistance through the regulation of BCL-2.
Recently, results of the differentially expressed miRNAs of tumors and matched normal tissues were deemed controversial, which is potentially due to the small sample sizes and variation in adopted techniques. Furthermore, the influence of other factors, such as sex, comorbidities (eg, immune status), social habits and age, on the expression level are still unknown. Overall, the alteration in the miRNA expression levels needs to be interpreted cautiously. Arroyo et al suggested that most circulating miRNAs existed in non-membrane-bound forms, consistent with ribonucleoprotein complexes, while only few existed in macrovesicles and exosomes.26 Exosomes are membrane-bound vesicles with diameters between 50 and 90 nm, which are generated from multivesicular bodies, and released from cells to circulation through the process of exocytosis.27 The exosomes are believed to participate in intercellular communication, since it was proposed that exosomes and Argonaute 2 (the effective component of the miRNA-induced silencing complex) can protect circulating miRNAs from degrading by RNase. In this study, we further validated tumor inhibition and promotion functions of miRNAs in exosomes, and found that it could be more suitable as a biomarker than circulating miRNAs in plasma. Multiple studies reported similar results. Bryant et al28 found that the expression levels of exosomal miR-141 and miR-375 in patients with metastatic PCa were higher than those in healthy controls, and suggested that they could be used as biomarkers for the diagnosis of PCa. Consistently, Li et al29 also found comparable results in their study. Huang et al30 demonstrated that high expression levels of serum exosomal miR-1290 and miR-375, negatively correlated with the survival rates of glucocorticoid-resistant PCa, suggesting that they could be used as biomarkers for the prognosis of such patients. Bhagirath et al31 suggested that the exosomal miR-1246 level was related to the pathological grade, positive transfer and poor prognosis of PCa, and could, therefore, be used as a biomarker for the diagnosis of PCa. Moreover, Mahn et al32 also found that the exosomal expression levels of miR-195 and let-7i were related to the Gleason Score and the postoperative positive margin, suggesting that they could be used to distinguish localized PCa and benign prostatic hyperplasia. Although the level of circulating miRNAs in exosomes is low, it has been proven that cancer cells significantly increase the circulating exosomes, suggesting that this increase in exosomal miRNA level is potentially tumor related. The interpretation of our study is limited, due to the small sample size and lack of analysis of cases after treatments. However, it can provide directions for further studies.
Data Sharing Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This work was supported by grant from China Postdoctoral Science Foundation (2019M650122).
Disclosure
The authors declare no conflicts of interest.
References
1. Wake N, Nussbaum JE, Elias MI, Nikas CV, Bjurlin MA. 3D printing, augmented reality, and virtual reality for the assessment and management of kidney and prostate cancer: a systematic review. Urology. 2020. doi:10.1016/j.urology.2020.03.066
2. Achard V, Bottero M, Rouzaud M, et al. Radiotherapy treatment volumes for oligorecurrent nodal prostate cancer: a systematic review. Acta Oncol. 2020:1–11. doi:10.1080/0284186X.2020.1775291.
3. McPhail S, Johnson S, Greenberg D, Peake M, Rous B. Stage at diagnosis and early mortality from cancer in England. Br J Cancer. 2015;112(Suppl S1):S108–115. doi:10.1038/bjc.2015.49
4. Bryan S, Masoud H, Weir HK, et al. Cancer in Canada: stage at diagnosis. Health Rep. 2018;29(12):21–25.
5. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
6. Begemann D, Wang Y, Yang W, Kyprianou N. Androgens modify therapeutic response to cabazitaxel in models of advanced prostate cancer. Prostate. 2020;80(12):926–937. doi:10.1002/pros.24015
7. Gourd E. New advances in prostate cancer screening and monitoring. Lancet Oncol. 2020;21(7):887. doi:10.1016/S1470-2045(20)30349-1
8. Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 1987;262:9412–9420.
9. Zamanian M, Fraser LM, Agbedanu PN, et al. Release of small RNA-containing exosome-like vesicles from the human filarial parasite brugia malayi. PLoS Negl Trop Dis. 2015;9(9):e0004069. doi:10.1371/journal.pntd.0004069
10. Denzer K, Kleijmeer MJ, Heijnen HF, Stoorvogel W, Geuze HJ. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J Cell Sci. 2000;113(Pt 19):3365–3374.
11. Liang B, Peng P, Chen S, et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J Proteomics. 2013;80:171–182. doi:10.1016/j.jprot.2012.12.029
12. Lasser C, Eldh M, Lotvall J. Isolation and characterization of RNA-containing exosomes. J Vis Exp. 2012;(59):e3037. doi:10.3791/3037
13. Mahmoodzadeh Hosseini H, Halabian R, Amin M, Imani Fooladi AA. Texosome-based drug delivery system for cancer therapy: from past to present. Cancer Biol Med. 2015;12:150–162. doi:10.7497/j.issn.2095-3941.2015.0045
14. Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–138. doi:10.1093/bioinformatics/btp612
15. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4. doi:10.7554/eLife.05005.
16. John B, Enright AJ, Aravin A, et al. Human microRNA targets. PLoS Biol. 2004;2(11):e363. doi:10.1371/journal.pbio.0020363
17. Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34(Web Server):W451–454. doi:10.1093/nar/gkl243
18. Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14. doi:10.1186/gb-2010-11-2-r14
19. Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007;36(Database):D480–484. doi:10.1093/nar/gkm882
20. Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi:10.1073/pnas.0804549105
21. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
22. Josson S, Gururajan M, Hu P, et al. miR-409-3p/-5p promotes tumorigenesis, epithelial-to-mesenchymal transition, and bone metastasis of human prostate cancer. Clin Cancer Res. 2014;20(17):4636–4646. doi:10.1158/1078-0432.CCR-14-0305
23. Sugatani T, Vacher J, Hruska KA. A microRNA expression signature of osteoclastogenesis. Blood. 2011;117(13):3648–3657. doi:10.1182/blood-2010-10-311415
24. Zhang HL, Qin X-J, Cao D-L, et al. An elevated serum miR-141 level in patients with bone-metastatic prostate cancer is correlated with more bone lesions. Asian J Androl. 2013;15(2):231–235. doi:10.1038/aja.2012.116
25. Corcoran C, Rani S, O’Driscoll L. miR-34a is an intracellular and exosomal predictive biomarker for response to docetaxel with clinical relevance to prostate cancer progression. Prostate. 2014;74(13):1320–1334. doi:10.1002/pros.22848
26. Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–5008. doi:10.1073/pnas.1019055108
27. Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16(4):415–421. doi:10.1016/j.ceb.2004.06.003
28. Bryant RJ, Pawlowski T, Catto JWF, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer. 2012;106(4):768–774. doi:10.1038/bjc.2011.595
29. Li Z, Ma YY, Wang J, et al. Exosomal microRNA-141 is upregulated in the serum of prostate cancer patients. Onco Targets Ther. 2016;9:139–148. doi:10.2147/OTT.S95565
30. Huang X, Yuan T, Liang M, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. 2015;67(1):33–41. doi:10.1016/j.eururo.2014.07.035
31. Bhagirath D, Yang TL, Bucay N, et al. microRNA-1246 is an exosomal biomarker for aggressive prostate cancer. Cancer Res. 2018;78(7):1833–1844. doi:10.1158/0008-5472.CAN-17-2069
32. Mahn R, Heukamp LC, Rogenhofer S, et al. Circulating microRNAs (miRNA) in serum of patients with prostate cancer. Urology. 2011;77(5):1265 e1269–1216. doi:10.1016/j.urology.2011.01.020
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.