Back to Journals » Drug Design, Development and Therapy » Volume 10
The interplay of post-translational modification and gene therapy
Authors Osamor V, Chinedu S , Azuh D, Iweala E, Ogunlana O
Received 7 January 2015
Accepted for publication 27 March 2015
Published 29 February 2016 Volume 2016:10 Pages 861—871
DOI https://doi.org/10.2147/DDDT.S80496
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Shu-Feng Zhou
Video abstract presented by Victor Chukwudi Osamor.
Views: 188
Victor Chukwudi Osamor,1–3 Shalom N Chinedu,3,4 Dominic E Azuh,3,5 Emeka Joshua Iweala,3,4 Olubanke Olujoke Ogunlana3,4
1Covenant University Bioinformatics Research (CUBRe) Unit, Department of Computer and Information Sciences, College of Science and Technology (CST), Covenant University, Ota, Ogun State, Nigeria; 2Institute of Informatics (Computational biology and Bioinformatics), Faculty of Mathematics, Informatics and Mechanics, University of Warsaw (Uniwersytet Warszawski), Warszawa, Poland; 3Covenant University Public Health and Well-being Research Group (CUPHWERG), Covenant University, 4Biochemistry and Molecular Biology Unit, Department of Biological Sciences, College of Science and Technology, Covenant University, Canaan Land, 5Department of Economics and Development Studies, Covenant University, Ota, Ogun State, Nigeria
Abstract: Several proteins interact either to activate or repress the expression of other genes during transcription. Based on the impact of these activities, the proteins can be classified into readers, modifier writers, and modifier erasers depending on whether histone marks are read, added, or removed, respectively, from a specific amino acid. Transcription is controlled by dynamic epigenetic marks with serious health implications in certain complex diseases, whose understanding may be useful in gene therapy. This work highlights traditional and current advances in post-translational modifications with relevance to gene therapy delivery. We report that enhanced understanding of epigenetic machinery provides clues to functional implication of certain genes/gene products and may facilitate transition toward revision of our clinical treatment procedure with effective fortification of gene therapy delivery.
Keywords: post-translational modification, gene therapy, epigenetics, histone, methylation
Introduction
The identical twins that originate from same egg and sperm are genetically identical, but may perhaps be different epigenetically due to influences that may occur later during zygotic cell differentiation and beyond. The epigenetics mechanism and its usefulness in the control of gene activity for several critical physiological and developmental processes, is sometimes marred by unattended process mistakes which become catastrophic when not urgently reversed by self or other cell processes. An intriguing question which the medics continue to grapple with or address is the understanding of the dynamics of heritable phenotypic changes without any underlying genotypic DNA sequence alterations.
Sometimes, our diagnosis fails and we are unable to pin-point any specific diagnostic basis for a continued ill-health condition. Cases like this may sometimes be an outcome of current limitation or gaps in modern medical delivery, and possibly an unnoticed epigenetic misregulation.
Post-translational modifications (PTM) in histone,1,2 DNA methylation3,4 mechanisms, and gene therapy are emerging fields; hence details of their processes need to be rigorously explored. Invariably, chromatin remodeling, which is an epigenetic modification, requires a great deal of ATP to form complexes which tend to shift and alter the stability of the nucleosome. In addition, there exists some histone variants which alter the functional mechanism of the nucleosome thereby creating epigenetic occurrences. Nevertheless, the long noncoding RNAs and noncoding RNAs consisting of piRNAs and siRNAs respectively, are able to influence and direct the epigenetic machinery. This may perhaps be by recruitment of epigenetic enzymes to epigenetic sites in the genome.
These factors notwithstanding, environmental influences could impact and instigate epigenetic changes. Similarly, some chemicals function as obesogens and increase the likelihood of obesity in an individual, thereby causing diseases such as diabetes. Tributyltin is an organotin biocide with serious epigenetic impact in organisms, causing endocrine disorder when inhaled, absorbed through the skin, or ingested along with contaminated water or food.5 It is a general antifouling industrial agent for paints used for underwater ship hull. When the fetus is exposed to this chemical, it initiates the reprogramming of the fetal stem cells to preferentially differentiate into fat cells at the expense of bone cells by epigenesis, leading to a series of disorders and malformations.
The knowledge of gene products as a function of epigenetic change clearly marks the way to go toward gene therapy intervention. This article highlights the role of some proteins in epigenetics while noting some epigenetic drugs and examining special considerations in gene therapy techniques. In considering why DNA methylation may not be seen strictly as a true epigenetic occurrence, we follow up with an overview of nucleosome structure and histone marks and then unravel a compendium of recent advances and discoveries in PTMs. We finally look at future perspectives of synergy between epigenetics and gene therapy.
Epigenetic drug and gene therapy techniques
As great research results are born in epigenetic studies, gene therapy with personalized approach will revolutionize our current traditional treatment method of drug delivery. Gene therapy, which involves the targeted transfer of recombinant genes into ailing tissues affected by known or unknown changes in gene activity/expression, offers great hope in epigenetic landscape discovery. As noted in several studies,6,7 cancer is thought to be a disease principally attributed to epigenetic misregulation. Some authors believe that there are many causes of cancer, some of which are unknown, while the known ones include benzene, alcohol consumption, environmental poison, poisonous mushrooms, excessive sunlight exposure, genetic problems, obesity, viruses, radiation, etc.8,9 In addition, transposon10 insertion into DNA can also cause cancer, depending on the site of insertion.
As an output of epigenetics, epigenetic drugs can initiate a gene-repressive-oriented therapy administered to influence gene activity toward disease amelioration. Azacitidine injection at the correct dosage is shown to be effective in the treatment of several types of cancers, such as myelodysplastic syndromes, which are diseases of the bone marrow, including chronic myelomonocytic leukemia. Interestingly, the phase III randomized clinical trials of Fenaux et al11 indicate that Vidaza (Azacitidine) injection prolongs overall survival compared to conventional care regimens in elderly patients with low bone marrow blast count acute myeloid leukemia. Vidaza is a chemotherapeutic drug belonging to the antimetabolite class of drugs that represses the expression of genes responsible for the growth of cells. This ultimately blocks the growth of cancer by preventing the synthesis of DNA and RNA thereby inhibiting tumor cell growth in leukemia patients. The functional implications of certain genes and knowledge on how their products interact are important for improved clinical treatment procedure and gene therapy12 intervention.
As discussed in the next section on DNA methylation, the state of methylation of O6-methyl guanine DNA methyl transferase (MGMT) gene can define the impact of the success story of gene therapy by using a two-component vector, a transgene, and MGMT against tumor cells. Furthermore, the clinical implication is that temozolomide is known to be more effective in patients with brain tumors such as glioblastoma having methylated MGMT promoter. In nature, viruses are adapted with the ability to enter specific cells for the purpose of transferring genetic material borne by them as carrier vectors. This event can be tweaked experimentally in the lab by deleting or mutating sequences from the wild-type viruses that code for viral replication proteins. This is done in a way that they are substituted with expression cassettes that encode the gene of interest, rendering them unable to replicate in the host cell. This transformed wild-type virus, which is now called a recombinant virus/vector, still retains the ability to penetrate host cells, but while carrying a recombinant gene of interest for delivery at target tissues. In this way, several disease conditions are ameliorated through systemic circulation of a recombinant protein. An example is seen in hemophilia B where the adeno-associated virus (AAV) expression cassette is loaded with factor IX (FIX), which tends to stop hemorrhagic diathesis.13,14
Current challenges to a successful gene therapy revolve around:
- The antigen–antibody response of host cell to transduced gene.
- Proper selection of appropriate vector system to deliver the gene of interest, whose malfunctioning is responsible for the ailing condition.
- Consideration of a targeted tissue or organ, a critical aspect of any gene delivery strategy, which often affects the choice of vector system used.
- Size of recombinant gene, which should be small so as to be accommodated inside the vector to deliver it to the required site in the body. (This may pose a problem if it is too large to be packaged into the vector).
- The process of vector development and the entire gene therapy being laborious.
- Gene therapy treatment option being too expensive.
Popular targets of choice for the delivery of injected gene therapy vectors in clinical trials, for selected fit individuals, are liver and muscle.15,16 Upon administration, the natural immunity of the body may act, causing antibody binding and subsequent eviction of the introduced vector particle from the body.17 In this regard, the preferred choice of several investigators is the AAV. A unique feature of AAV is its use as a nonintegrating vector,18,19 principally due to the existence of an episome that facilitates a transient and attenuating effect20 in cell division processes in tissues such as the liver in some animals.21,22 This confers a wider acceptance for AAV as they are more readily accommodated by the host immune system compared to other types of viral vectors. Besides, the lentivirus subclass of the retroviruses, such as HIV, is an integrating virus that has high propensity to insert itself into the host DNA. They multiply continuously to yield several copies during replication, hence it has a longer lasting effect, especially when the immune system is evaded.
MeCP and MGMT in DNA methylation mechanisms
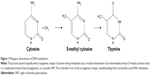
The methylation of DNA, unlike that of histones, may not be seen as a true epigenetic event or PTM due to the end product containing a sequence change. The underlying factor is that methylation epigenetic event occurs by the addition of methyl group to specific amino acid for histone methylation (PTM) or to the fifth carbon atom of the cytosine ring of a pyrimidine nucleotide (DNA methylation). It is known that this methylated cytosine quickly deaminates and transform to thymine (Figure 1), causing disorder. This complex case of disease exacerbation is seen as a complicated situation beyond the process of epigenetics, to possibly a single nucleotide polymorphism.
In 1994, Zhang and Mathews23 described an experiment that portrayed a 21-time higher susceptibility of methylated cytosine to deamination compared to unmethylated cytosine on the DNA. This suggests that DNA methylation is a hot spot for mutagenesis. Preferably, DNA methylation chose to occur mostly at locations of the CpG where there are either repeats or at transposon-inserted positions10 or regions. Although promoters have the tendency not to be methylated, due to the presence of CpG clusters of motif that is refractory to methylation,4 they are sometimes overwhelmed by hypermethylation in some human cancers that represses the activities of genes (tumor repressors gene) originally supposed to defend any methylation occurrence. With high susceptibility to deamination, Methyl-CpG (meCpG) in a CpG island creates room for the formation of a repressive chromatin structure. It occurs such that we have methylated CpG-binding proteins, MeCP1 and 2, which have great affinity binding to meCpG. MeCP proteins, with its inherent DNA-binding domain and transcription repression domain, can recruit other factors that condense the chromatin.
The purification and functional analysis of MeCP1 by Feng and Zhang3 reveals that it contains 10 other polypeptides, including MBD2 (the meCpG-binding protein), which recruit chromatin remodelers, deacetylate histones, resulting in gene silencing in methylated DNA. However, gel shift experiments with nuclear extracts and a CpG-methylated DNA probe indicate that recombinant MBD3L2 (meCpG-binding protein 3-like 2) can displace a form of the MeCP1 complex from methylated DNA to restore transcription.24,25
During DNA replication and as fallout of DNA methylation, in a cell with O6 methyl-guanine, the replication fork proceeds, thereby creating a mis-pair with thymine, thus leading to a GC→AT transition. A repair protein, MGMT is thereby activated to restore and preserve genome integrity. In this case, the MGMT protein accepts the methyl using its cysteine residue by a demethylation process of O6-methylguanine back to guanine. This role clinically portrays the importance of MGMT gene activities in ameliorating cases of patients susceptible to carcinoma and ultimately determines the success level of azacitidine, temozolomide, and gene therapy intervention.
Nucleosome and histone marks
Ever since the discovery of histone protein 1 and the winning of the 1910 Nobel Prize in Physiology or Medicine26 by Albrecht Kossel, other related discoveries continued to evolve. The idea that the number of chromosome is same in all somatic cells instigated Stedman and Stedman27 to suggest that histone totally represses gene expression.28 The landmark discovery by Allfrey et al29 opined that the chemical changes of methylation and acetylation impact the transcription process, and this revelation opened up several advances into today’s epigenetics. Rogers Kornberg, a 2006 Chemistry Nobel Laureate and American Scientist, unraveled the relationship between histones and DNA by discovering the nucleosome as repeating unit of histones along the DNA.30 The trio of Aaron Ciechanover, Avram Hershko, and Irwin Rose similarly won the 2004 Nobel Prize in Chemistry for their discovery of ubiquitin conjugation on lysine, which they reported as a procedure that picks protein for onward degradation.31
One of the most abundant and highly conserved eukaryotic proteins in the body are the histones, which are credited to facilitate the maintenance of structural framework and compaction of the genome. Indeed, their major role is more appreciated in the mediation of several biochemical processes32,33 that take place in the body at their N-terminal tail by other proteins.34,35 It is also known that not only proteins are able to recruit complexes that modify or remodels histones, but noncoding RNAs are also involved.36 In mouse, Pasque et al37 showed that X chromosome can be reprogrammed to maintain static irreversible process by inclusion of histone variant macroH2A.
The nucleosome represents a DNA wound almost 1.7 times around nuclear proteins called histones, which are of 4-paired types (H2A, H2B, H3, and H4) usually seen as an octamer core with a single unpaired H1 histone. The resulting architecture leaves H1 histone on the outside, thereby locking the entire set firmly as a tight unit as shown in Figure 2. It is now known that nucleosomes form a complex when one H1 histone interacts with another H1 histone. Earlier studies38–40 have shown that histones are able to recruit other proteins to form a complex.
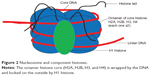  | Figure 2 Nucleosome and component histones. |
Each histone octamer bears a protruding tail with C-terminal end and a more reactive N-terminal that plays a pivotal role in epigenetic marks.
By this tendency of the 146 bp DNA length to wind on histones, nature has provided a mechanism for the 2 m DNA to be stacked in a 10 nm diameter nucleus. A collection of the chemical structure conformations of the resultant histone marks in the modification of amino acids like lysine, serine, and arginine are represented in Figure 3. N-monomethyl, NN-dimethyl, and NNN-trimethyl lysines are the histone methylation marks formed by the covalent modification of lysine via addition of one molecule of −CH3, two molecules of −CH3, and three molecules of −CH3 group, respectively. Another histone acetylation mark is created by the covalent addition of the −OCH3 group to lysine to form N-acetyl lysine. Similarly, arginine can covalently add one or two molecules of −CH3 group to form histone methylation marks such as N-monomethyl arginine and NN-dimethyl arginine, respectively. However, NN-dimethyl arginine has two types of conformation, namely, the symmetric and the asymmetric NN-dimethyl arginine. The PO4− (phosphate group) modifies the amino acid serine by phosphorylation to form phosphoserine.
Compendium of recent advances in PTM discoveries and their roles
Lysine acetylation
Acetylation of histone is important for transcription and DNA repairs. Acetyl-coenzyme A (CoA) is the major substrate for histone acetyl transferases (HATs). How the nucleus got the abundance of its acetyl CoA synthesized primarily in the mitochondria by the pyruvate dehydrogenase complex (PDC) remained a mystery until 2014, when Sutendra et al41 showed that mitochondrial PDC translocates to the nucleus at the instance of some signaling event to synthesize more acetyl CoA for continued histone acetylation. This was about the same time that Li et al42 discovered a novel family of histone acetylation readers called the YEATS domains, which is different from the usual royal family readers of bromodomains, chlorodomains or PHD. This difference shows that AF9 YEATS adopts an eight-stranded immunoglobin fold and utilizes a serine-lined aromatic “sandwiching” cage for acetyllysine readout. They showed that AF9 YEATS TF motif binds to H3K9ac readily than to other lysine acetylation marks like H3K18 and H3K27, while at the same time AF9 recruits DOTIL that ultimately deposits H3K79 methylation on active chromatin. This is arguably a positive count on the total number of possible cross talks between lysine acetylation and methylation.
Lysine methylation
Currently, interest on the group of proteins called the polycomb has grown, especially to explore the biological role of PRC2 complexes that contains Ezh2/E(Z) protein component.43 This is because Ezh2/E(Z) is able to engage in the methylation of H3 at lysine K27 to yield a histone mark (H3K27me1) indicted for heterochromatin formation (closed chromatin that represses gene expression). Similarly, further studies reveal that at histone mark H3 lysine 9 position, Eed/ESC which is another member of PRC2 complex, is able to engage a methylated H3K27me3 by binding to it44 and using its domains to interact with itself,45 thereby stimulating methyltransferase for Ezh2.46,28
Grossniklaus and Paro47 report that polycomb genes are for silenced chromatin development, and they participate in multimeric complexes where they bind to specific histone modification such as H3K27me3 and H2AK119ub1 to repress transcription processes. The Suv39 H1 (suppressor of variegation 3–9 homologue 1 [Drosophila] gene codes for histone methyl transferase [HMT]) responsible for trimethylating lysine 9 of histone 3 (H3) – H3K9me3. Bannister et al48 reported that a methylated histone H3 at position 9 lysine led to the recruitment of heterochromatin protein (HP1), which is a transcription repressor.49,50 This HP1 may perhaps choose to recruit Suv39H1 and in turn trigger the original methylation activity. Disease associated with Suv39H1 is idiopathic pulmonary fibrosis. This is similar to Suv91_Human that encodes histone trimethyltransferase, which specifically uses monomethylated H3K9 (H3K9me1) to trimethylate in other to yield H3K9me3 mark. This again suggests that several molecular activities may occur in a revolving cyclic manner via interactions of same or different protein component for the same histone mark. Ultimately, the effect of these repeated interactions by several different domains is quite complicated and will always have peculiar effects on the totality of the organism’s constitution and life.
Phosphorylation
Phosphorylation, which adds PO4− via kinases to serine or threonine, applies not only to H2A and H2B but also H3 and H4 histones. Phosphorylation is known to be reversed by phosphatases like PPI, and phosphorylation has functional roles in transcription, mitosis, DNA repair, and apoptosis. Specific examples include H2BS33ph (fruit fly) for DNA repair and condensation, H3S10ph supports apoptosis and H4S1ph for DNA repair and chromatin assembly. More recently, the knowledge horizon has been expanded by the work of Basnet et al51 who demonstrated that transcription elongation is impaired by tyrosine 57 mutation in H2A (Y57F) or casein kinase 2 (CK2) inhibition in both yeast and mammals. Furthermore, the strange mutation is implicated in the concomitant loss of active transcription in H3K4me3 and H3K 79me3, and another loss of H2B mono-ubiquitination in Spt-Ada-Gcn5 acetyltransferase (SAGA) complex portrays that the critical role of SAGA is phosphorylation dependent.
Sometimes, PTM-associated proteins exhibit the characteristics of allosteric regulation by metabolites around their binding site, but not attaching at the actual active/binding site. This depicts a usual conserved regulatory mechanisms phenomenon. Some phosphorous-containing PTM such as the primary glycolytic electrolytic intermediate 1,3-bisphosphoglycerate are capable of reacting and modifying specific lysine under no required catalytic conditions to produce 3-phosphoglyceryl-lysine (pgK), which ultimately inhibits glycolytic activities.52
Ubiquitination
Unlike acetylation and methylation, which covalently add acetyl and methyl groups, respectively, to histone protein such as lysine, ubiquitin modifies lysine residues by an interesting process of conserved conjugation cascade.53,54 Ubiquitin is common in eukaryotes and is involved in several body processes. However, they look similar but have different sequences with a signature diglycine sequence that is exposed after proteolytic processing. They are characterized by a biochemical mechanism of isopeptide bond that is formed between the modifier’s terminal glycine and an amino group of the target protein.
Ubiquitin has a wide variety of substrates, but most of its various domains are highly specific. Examples of its domains are activating enzymes E1, conjugating enzyme E2, and RING E3 domains. Ubiquitin’s modification of lysine will first involve the conjugative cascade machinery involving the domains that recognized the substrate lysine. It is important to state that Mdm2 is a RING E3 domain that controls the level of the tumor suppressor p53 interactions.55 The binding of Mdm2 with a hydrophobic cleft to p53 with a hydrophobic face interrupts the transcriptional activities of p53, which in turn induces Mdm2 to aid its nuclear transport. This is a typical case of autoregulatory negative feedback mechanism.56
SUMOylation
SUMOylation is the covalent addition of small ubiquitin-like modifiers to proteins by which the lysine at specific positions in a histone are marked for critical biological roles. These roles include protein homeostasis, trafficking and signal transduction, protein localization, gene regulation, and nucleocytoplasmic signaling and transport, are subject to reversible sumoylation, and are beginning to attract attention as an intervention method against diseases.57 The tumor suppressor protein, INhibitor of Growth (ING) exists in five isoforms (ING1–5), and they repress cell growth when overexpressed. Satpathy et al58 found that lysine K193 is a good ING1b SUMO acceptor site. This is believed to activate ING1b SUMOylation on K193, which is facilitated by S199D phosphomimic mutant. Ultimately, the binding of ING1b to promoter ISG15 and DGCR8 regulates transcription.
Proline isomerization
Through peptidyl-prolyl (proline) isomerase (PPIases) catalysis, the amino acid proline has a unique attribute of undergoing isomerism by its ability to interconvert between the cis and trans isomers (Figure 4) of the N-terminal amide bond.59 Conformations of these isomers have a role to play in protein folding and molecular switch formation in metabolic pathways for several processes, and reactions in this class of molecules are usually slow due to the double bond but are usually catalyzed by peptidyl-prolyl isomerase. Camilloni et al60 showed that a proline isomerase called cyclophilin A, “uses electrostatic handle” mechanism by creating:
Electrostatic environment in its catalytic site that rotates a peptide bond in the substrate by pulling the electric dipole associated with the carbonyl group preceding the peptide bond itself.60
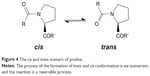  | Figure 4 The cis and trans isomers of proline. |
This outcome suggests a situation for the application of electrostatics in explaining enzyme catalytic process.
Different states like stress and several metabolic signals may affect and influence gene transcription. The organism has to respond to these influences, and changes will be noticed in its gene transcription profile to ensure that the organism survives or grows at the optimum prevailing state. Howe et al61 demonstrated that H3K14ac is responsible for change of state and the cis–trans isomerization of H3 at alanine-15-proline-16 peptide bond. Changes in lysine 14 (K14) acetylation improve the availability of the conformation of A15-P16trans. The ribosomal genes use K4me3 for repression during stress and are affected by the balancing action of Spp1 subunit of Set1 K4 methyltransferase complex and Jhd2 demethylases.
Biotinylation
The addition of biotin to proteins or other substances, a phenomenon often called biotinylation has found importance in application areas such as tagging and labeling of biomolecules. Due to biotin’s tight coupling with streptavidin, it is also known to efficiently refract light or fluoresces under light to reveal tags placed on them. However, there are various methods of biotinylation that range from chemical methods to enzymatic methods using some bacteria. In vivo biotinylation of bacterial magnetic particles (BacMPs) from Magnetospirillum magneticum AMB-1 was carried out by Maeda et al62 using simultaneous expression method of biotin acceptor protein (BAP) fused with Mms13 (which is BacMP surface protein) in truncated, DNA-binding domain-free E. coli biotin ligase. This biotinylation was confirmed by applying the resultant biotinylated BAP-BacMP to alkaline phosphatase-conjugated anti-biotin antibody and exposing it to streptavidin which has a higher affinity for in vivo biotinylation compared to in vitro biotinylation.
Biotinylation of RNA adds biotin to lysine protein molecules during investigative studies on RNA–protein interaction or in protein–protein interaction or DNA–protein interaction. Some nucleotides are also biotinylated. All these interactions with biotin are able to invoke some PTM to occur, including chromatin remodeling activities as demonstrated in E. coli with SW1/SNF2 proteins for RNA-directed binding.63
ADP-ribosylation
These sets of PTM are not well characterized relatively compared to other PTM. This may be attributable to the less frequent nature of their activities, as summed by the fact that they were discovered later. ADP-ribosyltransferase64 facilitates the addition of one or more moieties of ADP-ribose (comparable to methyl or acetyl) from nicotinamide adenine dinucleotide (NAD+) to the glutamate (E) or aspartic acid or arginine it modifies. This has found application in DNA repair and toxicology, where microbes like M. cholerae65 releases proteinous ADP-ribosylate-based toxins that act against human cells by adding moieties responsible for the deadly diarrhea.
Citrullination
Citrullination or deimination is the conversion of an arginine residue to the noncoded amino acid citrulline, mediated by peptidylarginine deiminases (PADIs). This enzyme has been linked with autoimmunity, cancer, neurodegenerative disorders, prion diseases, and thrombosis. In a recent work, Christophorou et al66 identified a linker histone (H1) variant that aids compact chromatin. However, H1 variant displacement from chromatin on citrullination of arginine activates transcription and may influence pluripotency.
Newer lysine modifications
Newer protein lysine modifications include butyrylation,67 propionylation,68 malonylation,69,70 crotonylation,71 and succinylation.72 Currently, there is the provision of a compendium of protein lysine modification database (CPLM) updated from a previously developed compendium of protein lysine acetylation (CPLA). The authors revealed that manual collection of experimentally identified substrate and sites was carried out for 12 types of protein lysine modifications to attain a current capacity of 203,972 modification events on 189,919 modified lysines in 45,748 proteins for 122 species.73 This suggests an increase in cross talk activities, which is estimated to be highest between acetylation and methylation as opposed to other PTMs.
Future perspective of epigenetics in gene therapy advances
It becomes imperative that as far as antibody binding response is a challenge, appropriate choice of vector and targeted tissue/organ consideration becomes inevitable. Since in some cases, there could be possible avoidance of systemic circulation (in vivo) by alternative means without necessarily injecting the patient with vector, strategies for transduction by direct application on the tissue (ex vivo).12 This suggests that perhaps the best way to advance is to re-emphasize research focus on direct application. However, it is impossible, and may be impracticable, to isolate some tissue/organ for such treatment delivery. Perhaps, evolution of more methods is desirable in this direction to ensure that vector particles are delivered to the target sites unhindered.
In addition to the impending challenges, there is the difficulty in packaging some relatively large genes/products of interest into vector particles due to their large size. This is known to hinder some treatment advances in hemophilia A, where the relatively large size of human factor VIII (FVIII) is a challenge for packaging into AAV2. Notwithstanding, McIntosh et al74 was able to obtain some level of successes while contending with difficulties inherent in the study of FVIII in mouse and nonhuman primates. This suggests that McIntosh et al’s codon optimized human FVIII variant (codop-hFVIII-V3) is safe and potent, at least in nonhuman animals, raising great hope that a gene therapy solution for hemophilia A in humans is at sight.
The process of generating and manufacturing a viral vector particle like AAV remains a tedious process further surmounted by a complex cellular procedure. This is evident in the work of Allay et al75 and Nathwani et al14 despite the success story of hemophilia B gene therapy. Gene therapy is a laborious relatively complex process with a risky nature compared to the traditional way of drug administration or conventional treatment regimen taken either orally, intramuscularly, or intravenously. Due to the attendant difficulties, gene therapy is currently too expensive for developing nations. The hope is that as more discoveries and improvements are made, especially in the epigenetics domain, it will in turn ignite gene therapy advances, which will necessitate a drop in price similar to what we witnessed in next-generation sequencing evolution.
Across the various modifications, are some antagonistic situations of feedback mechanisms which can be investigated and exploited to further advance gene treatment delivery. Mdm2, which is a RING E3 domain in ubiquitination, is overexpressed in several human cancers, and the inhibition of Mdm2 expression has been shown to lead to the activation of p53.76 When the binding of Mdm2/p53 is disrupted, p53 is activated; so a suitable small-molecule antagonist is needed to bind Mdm2 into the binding pocket of p53 to prevent the its ubiquitination and thereby opening of its pathway.22,56 This suggests that cancers arising from this type of modification, especially those indicted via the active p53 pathway, may be arrested by using therapy along the lines of this knowledge.
Originally, the widely used retroviral episomal transfer frequently cause biosafety77 issues and concerns in gene therapy delivery, resulting in several clinical trial failures.78,79 This is principally caused by the integration of the vector genome into the target cell chromatin thereby deregulating neighboring genes subsequently by the phenomenon of insertional mutagenesis. Recent advances has led to improved mutation of the retroviral integrase that converts these vectors into safer, transient, and integration-deficient gene delivery vehicles for gene therapy, thus reducing integration frequencies by 100- to 1,000-fold via introduction of a missense mutation into the DDE catalytic triad of retroviral integrase.80 Furthermore, Schott et al81 recently provided a proof of concept for episomal transfer of transcription factor Oct4, which was potent enough to mediate conversion of human fibroblasts stably expressing Klf4, Sox2, and c-Myc into induced pluripotent stem cells, which were mainly free of residual Oct4 vector integration. Interestingly, this evidence further confirms that the use of episomal expression enhanced by epigenetic modifiers as Oct4 activity increases following fusion to a minimal transactivation motif of herpes simplex virus VP16. In addition, long terminal repeat-driven γ-retroviral vectors’ architecture is seen as the most suitable vector for episome transfer of transcription factors for cell fate conversion, which further suggests the provision of new insight into a safer gene therapy treatment delivery.
In epigenetics, there exists a relationship between the effect of DNA methylation and the choice of vector for gene therapy. It has been discovered that self-complementary AAV vectors stimulate toll-like receptor 9, with stimulation stronger than that which occurs with single-stranded AAV vectors.82,83 This is because toll-like receptor 9 is a well-known pattern recognition receptor able to recognize unmethylated CpG dinucleotides. It is not clearly understood how the stimulation of toll-like receptor 9 will impact on vector’s efficacy and design strategies, and, as such, this has become an area to be explored.84,85 So far, after several decades of research, there are some rays of successes, but with greater hope that epigenetics research output will be the final knowledge depot for gene therapy advances in quest for healthy mankind.
Conclusion
The pivotal task in ameliorating epigenetic misregulation is to administer products with inherent ability to interact either directly or indirectly with the misregulated gene to stabilize or suppress its expression. The indirect approach may require the induction of other known repressors proteins complexes. Gene therapy has shown to handle some cases of absence or lack of specific gene activity, and the administration of such genes through recombinant vectors has produced some results. As molecular research effort progresses with time, more information have continued to emerge, giving further insight on new mechanisms and sources of epigenetic modifications, especially in humans. Over the past few years, minor epigenetic misregulation has been seen as synonymous with disease conditions like cancer, despite its positive role in developmental processes. Nevertheless, the enhanced understanding of epigenetic machinery will always provide clues on the functional implications of certain genes and may perhaps draw us close from different dimensions toward revision of our clinical treatment procedure with efficient infusion of gene therapy.12 Our increasing knowledge of the role of the MGMT gene and similar genes in combination with epigenetic drugs can fortify our advancement and further build on the gains of gene therapy in ameliorating cases of complex diseases.
Acknowledgments
This collaborative work was carried out during the tenure of Victor C Osamor on an ERCIM “Alain Bensoussan”/Marie Curie Fellowship at University of Warsaw, Poland. The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7/2007–2013) under grant agreement number 246016. We are grateful to Prof Jerzy Tiuryn, Mr D Adamiak, Prof Pawel Strzelecki, Prof Andrzej Tarlecki, Małgorzata Koślacz, and Małgorzata Smoderek for their support. Others include Mateusz Lacki, Shamba Sankar Mondal, Jaroslaw Paszek, Michal Wozniak, Pawel Bednarz, Agnieszka Mykowiecka, Aleksander Jankowski, Kyzysztof Gogolewski, Julia Herman-Izycka, Piotr Dittwald, Maciek Sykulski, Ilona Grabowicz, Rafal Zaborowski, Michal Startek, Pawel Gorecki, Bartek Wilczynski, and Anna Gambin.
Disclosure
The authors report no conflicts of interest in this work.
References
Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. | ||
Erkmann J. Histone modification research methods. Mater Methods. 2011;1:92. | ||
Feng Q, Zhang Y. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 2001;15:827–832. | ||
Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. | ||
Antizar-Ladislao B. Environmental levels, toxicity and human exposure to tributyltin (TBT)-contaminated marine environment: a review. Environ Int. 2008;34:292–308. | ||
Bernstein BE, Kamal M, Lindblad-Toh K, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. | ||
Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. | ||
Wolffe AP, Matzke MA. Epigenetics: regulation through repression. Science. 1999;286:481–486. | ||
Jauhari S, Rizvi SAM. Mining gene expression data focusing cancer therapeutics: a digest. IEEE/ACM Trans Comput Biol Bioinform. 2014;11:533–547. | ||
Startek M, Le Rouzic A, Capy P, Grzebelus D, Gambin A. Genomic parasites or symbionts? Modeling the effects of environmental pressure on transposition activity in asexual populations. Theor Popul Biol. 2013;90:145–151. | ||
Fenaux P, Mufti GJ, Hellström-Lindberg E, et al. Azacitidine prolongs overall survival compared with conventional care regimens in elderly patients with low bone marrow blast count acute myeloid leukemia. J Clin Oncol. 2010;28:562–569. | ||
Zhang L, Thrasher AJ, Gaspar HB. Current progress on gene therapy for primary immunodeficiencies. Gene Ther. 2013;20:963–969. | ||
Cancio MI, Reiss UM, Nathwani AC, Davidoff AM, Gray JT. Developments in the treatment of hemophilia B: focus on emerging gene therapy. Appl Clin Genet. 2013;6:91–101. | ||
Nathwani AC, Rosales C, McIntosh J, et al. Long-term safety and efficacy following systemic administration of a self-complementary AAV vector encoding human FIX pseudotyped with serotype 5 and 8 capsid proteins. Mol Ther. 2011;19:876–885. | ||
Manno CS, Chew AJ, Hutchison S, et al. AAV-mediated factor IX gene transfer to skeletal muscle in patients with severe hemophilia B. Blood. 2003;101:2963–2972. | ||
Buchlis G, Podsakoff GM, Radu A, et al. Factor IX expression in skeletal muscle of a severe hemophilia B patient 10 years after AAV-mediated gene transfer. Blood. 2012;119:3038–3041. | ||
Manno CS, Pierce GF, Arruda VR, et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med. 2006;12:342–347. | ||
Davidoff AM, Gray JT, Ng CYC, et al. Comparison of the ability of adeno-associated viral vectors pseudotyped with serotype 2, 5, and 8 capsid proteins to mediate efficient transduction of the liver in murine and nonhuman primate models. Mol Ther. 2005;11:875–888. | ||
Hurlbut GD, Ziegler RJ, Nietupski JB, et al. Preexisting immunity and low expression in primates highlight translational challenges for liver-directed AAV8-mediated gene therapy. Mol Ther. 2010;18:1983–1994. | ||
Varnavski AN, Zhang Y, Schnell M, et al. Preexisting immunity to adenovirus in rhesus monkeys fails to prevent vector-induced toxicity. J Virol. 2002;76:5711–5719. | ||
Suzuki M, Bertin TK, Rogers GL, et al. Differential type I interferon-dependent transgene silencing of helper-dependent adenoviral vs adeno-associated viral vectors in vivo. Mol Ther. 2013;21: 796–805. | ||
Varnavski AN, Calcedo R, Bove M, Gao G, Wilson JM. Evaluation of toxicity from high-dose systemic administration of recombinant adenovirus vector in vector-naïve and pre-immunized mice. Gene Ther. 2005;12:427–436. | ||
Zhang X, Mathews CK. Effect of DNA cytosine methylation upon deamination-induced mutagenesis in a natural target sequence in duplex DNA. J Biol Chem. 1994;269:7066–7069. | ||
Jin S-G, Jiang C-L, Rauch T, Li H, Pfeifer GP. MBD3L2 interacts with MBD3 and components of the NuRD complex and can oppose MBD2-MeCP1-mediated methylation silencing. J Biol Chem. 2005;280:12700–12709. | ||
Fujikane R, Sanada M, Sekiguchi M, Hidaka M. The identification of a novel gene, MAPO2, that is involved in the induction of apoptosis triggered by O6-methylguanine. PLoS One. 2012;7:e44817. | ||
Ramsay SW, Rutherford E, Curie MS, Soddy F, et al. Nobel Lectures Including Presentation Speeches and Laureates’ Biographies: Chemistry 1901–1921. Amsterdam, the Netherlands: Elsevier; 1966. | ||
Stedman E, Stedman E. Cell specificity of histones. Nature. 1950;166:780–781. | ||
Felsenfeld G. A brief history of epigenetics. Cold Spring Harb Perspect Biol. 2014;6:a018200. | ||
Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci U S A. 1964;51:786–794. | ||
Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. | ||
Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle (Nobel Lecture). Cell Death Differ. 2005;12:1191–1197. | ||
Wallis JW, Hereford L, Grunstein M. Histone H2B genes of yeast encode two different proteins. Cell. 1980;22:799–805. | ||
Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A. Structure of the nucleosome core particle at 7 Å resolution. Nature. 1984;311:532–537. | ||
Durrin LK, Mann RK, Kayne PS, Grunstein M. Yeast histone H4 N-terminal sequence is required for promoter activation in vivo. Cell. 1991;65:1023–1031. | ||
Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. | ||
Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP). J Vis Exp. 2012;25:3912. | ||
Pasque V, Halley-Stott RP, Gillich A, Garrett N, Gurdon JB. Epigenetic stability of repressed states involving the histone variant macroH2A revealed by nuclear transfer to Xenopus oocytes. Nucleus. 2011;2:533–539. | ||
Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. | ||
Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002;111:771–778. | ||
Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. | ||
Sutendra G, Kinnaird A, Dromparis P, et al. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell. 2014;158:84–97. | ||
Li Y, Wen H, Xi Y, et al. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell. 2014;159: 558–571. | ||
Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. | ||
Hansen KH, Bracken AP, Pasini D, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. | ||
Margueron R, Li G, Sarma K, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. | ||
Margueron R, Justin N, Ohno K, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. | ||
Grossniklaus U, Paro R. Transcriptional silencing by polycomb-group proteins. Cold Spring Harb Perspect Biol. 2014;6:a019331. | ||
Bannister AJ, Zegerman P, Partridge JF, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. | ||
Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. | ||
Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. | ||
Basnet H, Su XB, Tan Y, et al. Tyrosine phosphorylation of histone H2A by CK2 regulates transcriptional elongation. Nature. 2014;516(7530):267–271. | ||
Moellering RE, Cravatt BF. Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science. 2013;341:549–553. | ||
Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. | ||
Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–1107. | ||
Yang Y, Li C-CH, Weissman AM. Regulating the p53 system through ubiquitination. Oncogene. 2004;23:2096–2106. | ||
Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. | ||
Flotho A, Melchor F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82(1):357–385. | ||
Satpathy S, Guérillon C, Kim T-S, et al. SUMOylation of the ING1b tumor suppressor regulates gene transcription. Carcinogenesis. 2014;35:2214–2223. | ||
Fischer S, Dunbrack RL, Karplus M. Cis-trans imide isomerization of the proline dipeptide. J Am Chem Soc. 1994;116:11931–11937. | ||
Camilloni C, Sahakyan AB, Holliday MJ, et al. Cyclophilin A catalyzes proline isomerization by an electrostatic handle mechanism. Proc Natl Acad Sci U S A. 2014;111:10203–10208. | ||
Howe FS, Boubriak I, Sale MJ, et al. Lysine acetylation controls local protein conformation by influencing proline isomerization. Mol Cell. 2014;55:733–744. | ||
Maeda Y, Yoshino T, Matsunaga T. In vivo biotinylation of bacterial magnetic particles by a truncated form of Escherichia coli biotin ligase and biotin acceptor peptide. Appl Environ Microbiol. 2010;76:5785–5790. | ||
McKinley BA, Sukhodolets MV. Escherichia coli RNA polymerase-associated SWI/SNF protein RapA: evidence for RNA-directed binding and remodeling activity. Nucleic Acids Res. 2007;35:7044–7060. | ||
Belenky P, Bogan KL, Brenner C. NAD+ metabolism in health and disease. Trends Biochem Sci. 2007;32:12–19. | ||
De Haan L, Hirst TR. Cholera toxin: a paradigm for multi-functional engagement of cellular mechanisms [Review]. Mol Membr Biol. 2004;21:77–92. | ||
Christophorou MA, Castelo-Branco G, Oliveira CS, et al. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. 2014;507:104–108. | ||
Chen Y, Sprung R, Tang Y, et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. 2007;6:812–819. | ||
Cheng Z, Tang Y, Chen Y, et al. Molecular characterization of propionyllysines in non-histone proteins. Mol Cell Proteomics. 2009;8:45–52. | ||
Peng C, Lu Z, Xie Z, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011;10:M111.012658. | ||
Xie Z, Dai J, Dai L, et al. Lysine succinylation and lysine malonylation in histones. Mol Cell Proteomics. 2012;11:100–107. | ||
Tan M, Luo H, Lee S, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. | ||
Zhang Z, Tan M, Xie Z, Dai L, Chen Y, Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol. 2011;7:58–63. | ||
Liu Z, Wang Y, Gao T, et al. CPLM: a database of protein lysine modifications. Nucleic Acids Res. 2014;42(Database issue):D531–D536. | ||
McIntosh J, Lenting PJ, Rosales C, et al. Therapeutic levels of FVIII following a single peripheral vein administration of rAAV vector encoding a novel human factor VIII variant. Blood. 2013;121: 3335–3344. | ||
Allay JA, Sleep S, Long S, et al. Good manufacturing practice production of self-complementary serotype 8 adeno-associated viral vector for a hemophilia B clinical trial. Hum Gene Ther. 2011;22: 595–604. | ||
Kussie PH, Gorina S, Marechal V, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. | ||
Hacein-Bey-Abina S, von Kalle C, Schmidt M, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348:255–256. | ||
Modlich U, Schambach A, Brugman MH, et al. Leukemia induction after a single retroviral vector insertion in Evi1 or Prdm16. Leukemia. 2008;22:1519–1528. | ||
Howe SJ, Mansour MR, Schwarzwaelder K, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. | ||
Wanisch K, Yáñez-Muñoz RJ. Integration-deficient lentiviral vectors: a slow coming of age. Mol Ther. 2009;17:1316–1332. | ||
Schott JW, Hoffmann D, Maetzig T, et al. Improved retroviral episome transfer of transcription factors enables sustained cell fate modification. Gene Ther. 2014;21:938–949. | ||
Rogers GL, Martino AT, Aslanidi GV, et al. Innate immune responses to AAV vectors. Front Microbiol. 2011;2:194. | ||
Martino AT, Suzuki M, Markusic DM, et al. The genome of self-complementary adeno-associated viral vectors increases toll-like receptor 9–dependent innate immune responses in the liver. Blood. 2011;117:6459–6468. | ||
Faust SM, Bell P, Cutler BJ, et al. CpG-depleted adeno-associated virus vectors evade immune detection. J Clin Invest. 2013;123:2994–3001. | ||
Martino AT, Basner-Tschakarjan E, Markusic DM, et al. Engineered AAV vector minimizes in vivo targeting of transduced hepatocytes by capsid-specific CD8+ T cells. Blood. 2013;121:2224–2233. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.