Back to Journals » Drug Design, Development and Therapy » Volume 10
The clinicopathological significance and ethnic difference of FHIT hypermethylation in non-small-cell lung carcinoma: a meta-analysis and literature review
Authors Wu X, Wu G, Yao X, Hou G, Jiang F
Received 22 March 2015
Accepted for publication 20 December 2015
Published 15 February 2016 Volume 2016:10 Pages 699—709
DOI https://doi.org/10.2147/DDDT.S85253
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Prof. Dr. Wei Duan
Xiaoyu Wu,1,* Guannan Wu,1,* Xuequan Yao,1 Gang Hou,2 Feng Jiang3
1Department of Surgical Oncology, Affiliated Hospital of Nanjing University of Traditional Chinese Medicine, Nanjing, 2Department of Respiratory Medicine, The First Hospital of China Medical University, Shenyang, 3Department of Thoracic Surgery, Jiangsu Cancer Hospital, The Affiliated Cancer Hospital of Nanjing Medical University, Nanjing, People’s Republic of China
*These authors contributed equally to this work
Abstract: Emerging evidence indicates that FHIT is a candidate tumor suppressor in many types of tumors including non-small-cell lung carcinoma (NSCLC). However, the prognostic value and correlation between FHIT hypermethylation and clinicopathological characteristics of NSCLC remains unclear. In this report, we performed a meta-analysis to evaluate the effects of FHIT hypermethylation on the incidence of NSCLC and clinicopathological characteristics of human NSCLC patients. Final analysis of 1,801 NSCLC patients from 18 eligible studies was performed. FHIT hypermethylation was found to be significantly higher in NSCLC than in normal lung tissue. The pooled odds ratio (OR) from ten studies included 819 NSCLC and 792 normal lung tissues (OR =7.51, 95% confidence interval [CI] =2.98–18.91, P<0.0001). Subgroup analysis based on ethnicity implied that FHIT hypermethylation level was higher in NSCLC tissues than in normal tissues in both Caucasians (P=0.02) and Asians (P<0.0001), indicating that the difference in Asians was much more significant. FHIT hypermethylation was also correlated with sex status, smoking status, as well as pathological types. In addition, patients with FHIT hypermethylation had a lower survival rate than those without (hazard ratio =1.73, 95% CI =1.10–2.71, P=0.02). The results of this meta-analysis suggest that FHIT hypermethylation is associated with an increased risk and poor survival in NSCLC patients. FHIT hypermethylation, which induces the inactivation of FHIT gene, plays an important role in the carcinogenesis and clinical outcome and may serve as a potential diagnostic marker and drug target of NSCLC.
Keywords: FHIT, methylation, tumor suppressor gene, meta-analysis, odds ratio, hazard ratio
Introduction
Lung cancers consist of two major histological types, non-small-cell lung carcinoma (NSCLC) and small-cell lung carcinoma. NSCLC consists of squamous cell carcinoma (SCC), adenocarcinoma (AD), large-cell carcinoma, and others. NSCLC accounts for ~85% of all lung cancers, and there are ~80% of NSCLC cases in advanced stage where the prognosis remains poor.1 Therefore, investigation of the mechanism of initiation, progression, and identification of prognostic markers is still needed for the selection of patients with NSCLC and to provide better individualized treatment. In recent years, a number of new tools, such as protein–protein interaction prediction approach,2 pathway data integration,3 and gene transcription analysis,4 were widely used in the study of epigenetic regulation and modification. Epigenetic modification of gene expression plays an important role in carcinogenesis. Epigenetic alterations, particularly aberrant DNA methylation, one of the best-characterized epigenetic modifications, contribute to tumor initiation and progression.5,6 Aberrant methylation of CpG dinucleotides is a commonly observed epigenetic modification in human cancer.5–7 Thus, the analysis of specific gene promoter methylation as a tool for diagnosis of tumors or its use as prognostic marker has been widely used for many different cancers including NSCLC.8
FHIT, also known as bis(5′-adenosyl)-triphosphatase, is one of the histidine triad gene family members and is an enzyme encoded by the FHIT gene.9,10 The FHIT gene locates the most common fragile site in the human genome, FRA3B (3p14.2), a region which frequently undergoes genomic rearrangement, biallelic loss, and cytogenetic abnormalities in tumors.9,11,12 Previous reports showed that FHIT was inactivated by the loss of heterozygosity and methylation in cancer cells, which indicated that FHIT is a tumor suppressor protein.13,14 Its precise function has been intensively studied in several tumors, by inducing cell cycle arrest and apoptosis, inhibition of cell proliferation, and increasing cell sensitivity to DNA-damaging agents.15–17 Lack of expression of FHIT protein by promoter methylation (hypermethylation) has been found to play an important role in lung alveolar differentiation regulation and epithelial tumorigenesis.18–21 Although previous studies indicated that inactivation of the FHIT is mainly induced by hypermethylation of FHIT gene, the reported rates of FHIT hypermethylation in NSCLC were remarkably diverse. Moreover, whether or not it is associated with the incidence and clinical characteristics of NSCLC is still unclear. The various results of these studies underpin the need for assessing the evidence of the relationship between FHIT inactivation and NSCLC. Hence, we conducted a systematic review and meta-analysis to quantitatively evaluate the effects of FHIT hypermethylation on the incidence and clinical characteristics of NSCLC.
Materials and methods
Search strategy and selection criteria
We searched PubMed, Embase, and ISI Web of Knowledge to identify studies from May 1, 1998 to October, 2015 using the search terms “lung” and “cancer or tumor or neoplasm or carcinoma”, “methylation”, and “FHIT or Fragile histidine triad protein or Bis (5′-adenosyl)-triphosphatase”. We also searched manually for the reference lists of the retrieved articles and reviews for additional articles.
Although our search did not have language limits initially, for the full-text reading and final evaluation, we only performed the review of the studies published in English language. After exclusion of nonrelevant and/or redundant publications from different databases, the remaining papers were evaluated in the full-text version for inclusion and exclusion criteria and for relevant articles in the reference lists. All searched data were retrieved. Authors’ bibliographies and references of selected studies were also searched for other relevant studies. The most complete study was chosen to avoid duplication if the same patient populations were reported in several publications.
Criteria that an eligible study had to meet were as follows: (1) FHIT hypermethylation evaluated in the primary NSCLC tissues, (2) research revealed the relationship between FHIT hypermethylation and NSCLC clinicopathological parameters and prognosis, (3) FHIT hypermethylation examined by polymerase chain reaction, and (4) studies provided sufficient information to estimate hazard ratio (HR) about overall survival (OS) and 95% confidence interval (CI). The exclusion criteria included the following: (1) letters, reviews, case reports, conference abstracts, editorials, and expert opinion, and (2) all publications regarding in vitro/ex vivo studies, cell lines, and human xenografts were also excluded.
Data extraction and methodological assessment
Two authors (XW and GW) independently reviewed and extracted data from eligible studies. Disagreements were resolved by discussion and consensus. Two authors (XY and GH) reviewed all the articles that fit inclusion and exclusion criteria. The following information was recorded for each study: the first author name, year of publication, sample source, number of cases, clinicopathological parameters, cancer tumor node metastasis stage, methylation detection method, methylation rate and/or expression, and follow-up. Data for study characteristics and clinical responses were summarized and organized into a table format. Heterogeneity of investigation was evaluated to determine whether or not the data of the various studies could be analyzed for a meta-analysis.
For the methodological evaluation of the studies, three investigators (XW, GW, and XY) read through each publication independently, and they assessed and scored them according to the REMARK guidelines and ELCWP quality scale.22,23 The three readers provided the quality scores and compared them, and then they reached a consensus value for each item.
Statistical analysis
Analysis was conducted using the STATA 12.0 (Stata Corporation, College Station, TX, USA) and Review Manager 5.2 (Cochrane Collaboration, Oxford, UK). The pooled frequency of FHIT hypermethylation and 95% CIs were estimated. The frequency of FHIT hypermethylation was compared in different tumor characteristics. Heterogeneity among studies was evaluated with Cochran’s Q test24 and the I2 statistic.25,26 When heterogeneity was not an issue (I2 values <50%), a fixed-effect model was used to calculate parameters. If there was substantial heterogeneity (I2 values ≥50%), a random-effect model was used to pool data and attempt to identify potential sources of heterogeneity based on subgroup analyses. The pooled OR was estimated for the association between FHIT hypermethylation and clinicopathological features. P-values tailed <0.05 were considered statistically significant.
Publication bias was assessed by using a method reported by Egger et al.27 We also explored reasons for statistical heterogeneity using meta-regression, subgroup analysis, and sensitivity analysis. The analysis of meta-regression and publication bias was performed by using STATA version 10.0.
Results
Seventy publications were identified by the search method as described in the “Materials and methods” section. Fifty-two of those were excluded as they were laboratory studies, non-original articles (review), or studies irrelevant to the current analysis. Eventually, there were 18 studies from 2001 to 2014 included in final meta-analysis19,28–44 as shown in Figure 1. A total of 1,801 NSCLC patients from People’s Republic of China, South Korea, Japan, Egypt, Italy, and USA were enrolled. Their basic characteristics are summarized in Table 1.
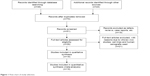  | Figure 1 Flow chart of study selection. |
We first determined that FHIT hypermethylation was significantly higher in NSCLC than in normal lung tissues. The pooled odds ratio (OR) from ten studies including 819 NSCLC and 792 normal lung tissues is shown in Figure 2 (OR =7.51, 95% CI =2.98–18.91, P<0.0001), indicating that FHIT hypermethylation plays an important role in the carcinogenesis of NSCLC. Subgroup analysis based on ethnicity implied that FHIT hypermethylation level was higher in NSCLC tissues than in normal tissues in both Caucasians (P=0.02) and Asians (P<0.0001) as shown in Figure 3, indicating that the difference in Asians was much more significant. Next, we determined whether or not FHIT hypermethylation rate was correlated with sex status. The pooled OR from eight studies included NSCLC from 742 males and 298 females, as shown in Figure 4 (OR =1.44, 95% CI =1.07–1.94, P=0.02), indicating that FHIT hypermethylation was correlated with sex status and it was higher in male than female. Then, we determined whether or not FHIT hypermethylation rate was correlated with smoking status. The pooled OR from ten studies including 287 and 818 NSCLC patients with and without smoking history is shown in Figure 5 (OR =0.74, 95% CI =0.55–1.00, P=0.05), indicating that FHIT hypermethylation was correlated with smoking status in NSCLC patients. We also determined whether or not FHIT hypermethylation was correlated with pathological types. The pooled OR from ten studies including 528 SCC and 527 AD patients is shown in Figure 6 (OR =1.49, 95% CI =1.15–1.93, P=0.003), which indicates that FHIT hypermethylation plays a more important role in the pathogenesis of SCC.
We analyzed 366 NSCLC patients pooled from three studies to assess whether or not the aberrant FHIT hypermethylation in NSCLC was associated with the differentiated status. As shown in Figure 7A, aberrant FHIT hypermethylation was not significantly higher in poorly differentiated NSCLC than that in moderately or highly differentiated NSCLC (OR =1.30, 95% CI =0.80–2.09, P=0.29). In addition, aberrant FHIT hypermethylation was also not significantly higher in advanced NSCLC (III and IV) than that in early-staged NSCLC (I and II) (OR =1.17, 95% CI =0.75–1.83, P=0.50; Figure 7B). These results suggest that FHIT hypermethylation may not play an important role in NSCLC progression and differentiation stages. There are four studies estimating the relationship between FHIT hypermethylation and OS in NSCLC patients. The pooled HR for OS showed that FHIT hypermethylation was associated with poor survival in NSCLC patients as shown in Figure 8 (HR =1.73, 95% CI =1.10–2.71, P=0.02).
A sensitivity analysis, in which one study was removed at a time, was conducted to assess the result stability. The pooled ORs and HRs were not significantly changed, indicating the stability of our analyses. The funnel plots were largely symmetric (Figure 9A–G), suggesting that there were no publication biases in the meta-analysis of FHIT hypermethylation and clinicopathological features.
Discussion
FHIT is genetically or epigenetically altered in many different kinds of primary and advanced carcinomas. Inactivation of FHIT by promoter hypermethylation plays an important role in tumorigenesis in several types of tumors including NSCLC.38,45–53 To date, there have been some studies describing the methylation status of FHIT in NSCLC; however, the roles of methylation of FHIT in NSCLC and clinical significance have not been thoroughly investigated. We conducted the meta-analysis to determine the correlation between FHIT hypermethylation and clinicopathological characteristics of NSCLC. Analysis of the pooled data showed that NSCLC had a higher hypermethylation than normal lung tissue, indicating that FHIT hypermethylation plays an important role in the carcinogenesis of NSCLC. Interestingly, subgroup analysis based on ethnicity implied that FHIT hypermethylation level was higher in NSCLC tissues than in normal tissues in both Caucasians (P=0.02) and Asians (P<0.0001) as shown in Figure 3, indicating that the difference in Asians was much more significant. Additional interesting findings included that FHIT hypermethylation was also correlated with sex status, smoking status, as well as pathological types. The results from the current study demonstrated that the hypermethylation rate of FHIT gene in NSCLC was significantly higher than that in the normal lung tissues, indicating that FHIT promoter hypermethylation was common in NSCLC. Therefore, detection of FHIT promoter hypermethylation may provide an invaluable diagnostic marker for NSCLC patients. Since changes in FHIT promoter hypermethylation are reversible, drug treatment through demethylation may be useful to delay carcinogenesis and progression and to improve prognosis. Lung cancer cell clones carrying conditional FHIT transgenes showed significant suppression of xenograft tumor growth after induction of expression of the FHIT transgene, suggesting that treatments to restore endogenous FHIT expression in lung cancers would result in decreased tumorigenicity.20 In addition, injection of 5-aza-2-deoxycytidine and trichostatin A in nude mice with established H1299 tumors showed suppressed growth of small tumors without apparent toxicity and responding tumors showed restoration of FHIT.20 These preclinical studies show the therapeutic potential of restoration of tumor suppressor expression through epigenetic modulation. This approach may bring new direction and hope for cancer treatment through gene-targeted therapy.
FHIT is thought to affect cellular function and behavior largely through its signaling properties. FHIT also activates caspase-8 and caspase-2, which causes the release of cytochrome c and finally induces apoptosis.54 FHIT and p53, the two most commonly altered tumor suppressor genes, might rely on common mediators and cross talk between these proteins in regulation of growth-related pathways; thus, the inactivation of both genes results in prominent deregulation of cell proliferation and tumor progression in lung cancer.55 A number of studies showed that inactivation of FHIT can cause tumor aberrant progression and link to clinicopathological characteristics.28,38,56–58 Therefore, FHIT can be considered as a tumor suppressor, and its inactivation could contribute to tumor progression and poor prognosis. Although only four studies evaluated the relationship between OS and FHIT hypermethylation in NSCLC, they showed very similar results.36,38,39,43 Based on this meta-analysis, the pooled HR for OS showed that FHIT hypermethylation was associated with poor survival in NSCLC patients (HR =1.73, 95% CI =1.10–2.71, P=0.02). Therefore, we may consider that FHIT hypermethylation in NSCLC tends to indicate a poor prognosis.
Consistent results were shown in sensitivity analyses, and no evidence of publication bias was found. This study has several potential limitations. First, the possibility of information and selection biases and unidentified confounders could not be completely excluded because all of the included studies were observational. Second, the searching strategy was restricted to articles published in English. Articles with potentially high-quality data that were published in other languages were not included because of anticipated difficulties in obtaining accurate medical translation. Hence, caution should be taken when our findings are interpreted among the general population.
In conclusion, our meta-analysis showed that NSCLC tissue had a higher FHIT hypermethylation than normal lung tissue, higher in male than female, higher in nonsmoker than smoker, and higher in SCC than AD. In addition, FHIT hypermethylation is associated with an increased risk and poor survival in NSCLC. FHIT hypermethylation, which induces the inactivation of FHIT gene, may play an important role in the carcinogenesis and clinical outcome and may serve as a potential diagnostic marker and drug target of NSCLC. Further large-scale studies, especially multicenter and well-matched cohort research, will provide more insight into the role of FHIT in the prognosis and clinical implementation of NSCLC patients.
Acknowledgments
This study was supported by National Science Foundation of China (81402523 to XW, 81472702 to FJ), Jiangsu Province “Three three three” high peak (BRA2015491 to FJ), and the Natural Science Foundation of Jiangsu Province (BK2012482 to FJ).
Disclosure
The authors report no conflicts of interest in this work.
References
Ramalingam S, Belani C. Systemic chemotherapy for advanced non-small cell lung cancer: recent advances and future directions. Oncologist. 2008;13 Suppl 1:5–13. | ||
Zhou H, Gao S, Nguyen NN, et al. Stringent homology-based prediction of H. sapiens-M. tuberculosis H37Rv protein-protein interactions. Biol Direct. 2014;9:5. | ||
Zhou H, Jin J, Zhang H, Yi B, Wozniak M, Wong L. IntPath – an integrated pathway gene relationship database for model organisms and important pathogens. BMC Syst Biol. 2012;6 Suppl 2:S2. | ||
Portal D, Zhou H, Zhao B, et al. Epstein-Barr virus nuclear antigen leader protein localizes to promoters and enhancers with cell transcription factors and EBNA2. Proc Natl Acad Sci U S A. 2013;110:18537–18542. | ||
Delpu Y, Cordelier P, Cho WC, Torrisani J. DNA methylation and cancer diagnosis. Int J Mol Sci. 2013;14:15029–15058. | ||
Ma X, Wang YW, Zhang MQ, Gazdar AF. DNA methylation data analysis and its application to cancer research. Epigenomics. 2013;5:301–316. | ||
Ghavifekr Fakhr M, Farshdousti Hagh M, Shanehbandi D, Baradaran B. DNA methylation pattern as important epigenetic criterion in cancer. Genet Res Int. 2013;2013:317569. | ||
Fleischhacker M, Dietrich D, Liebenberg V, Field JK, Schmidt B. The role of DNA methylation as biomarkers in the clinical management of lung cancer. Expert Rev Respir Med. 2013;7:363–383. | ||
Ohta M, Inoue H, Cotticelli MG, et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587–597. | ||
Pekarsky Y, Campiglio M, Siprashvili Z, et al. Nitrilase and Fhit homologs are encoded as fusion proteins in Drosophila melanogaster and Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:8744–8749. | ||
Romero I, Martinez M, Garrido C, et al. The tumour suppressor Fhit positively regulates MHC class I expression on cancer cells. J Pathol. 2012;227:367–379. | ||
Pichiorri F, Palumbo T, Suh SS, et al. Fhit tumor suppressor: guardian of the preneoplastic genome. Future Oncol. 2008;4:815–824. | ||
Hassan MI, Naiyer A, Ahmad F. Fragile histidine triad protein: structure, function, and its association with tumorogenesis. J Cancer Res Clin Oncol. 2010;136:333–350. | ||
Pekarsky Y, Palamarchuk A, Huebner K, Croce CM. FHIT as tumor suppressor: mechanisms and therapeutic opportunities. Cancer Biol Ther. 2002;1:232–236. | ||
Huang Q, Liu Z, Xie F, et al. Fragile histidine triad (FHIT) suppresses proliferation and promotes apoptosis in cholangiocarcinoma cells by blocking PI3K-Akt pathway. ScientificWorldJournal. 2014;2014:179698. | ||
Rimessi A, Marchi S, Fotino C, et al. Intramitochondrial calcium regulation by the FHIT gene product sensitizes to apoptosis. Proc Natl Acad Sci U S A. 2009;106:12753–12758. | ||
Trapasso F, Pichiorri F, Gaspari M, et al. Fhit interaction with ferredoxin reductase triggers generation of reactive oxygen species and apoptosis of cancer cells. J Biol Chem. 2008;283:13736–13744. | ||
Tan S, Sun C, Wei X, et al. Quantitative assessment of lung cancer associated with genes methylation in the peripheral blood. Exp Lung Res. 2013;39:182–190. | ||
Verri C, Roz L, Conte D, et al. Fragile histidine triad gene inactivation in lung cancer: the European Early Lung Cancer project. Am J Respir Crit Care Med. 2009;179:396–401. | ||
Cantor JP, Iliopoulos D, Rao AS, et al. Epigenetic modulation of endogenous tumor suppressor expression in lung cancer xenografts suppresses tumorigenicity. Int J Cancer. 2007;120:24–31. | ||
Han SY, Iliopoulos D, Druck T, et al. CpG methylation in the Fhit regulatory region: relation to Fhit expression in murine tumors. Oncogene. 2004;23:3990–3998. | ||
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst. 2005;97:1180–1184. | ||
Steels E, Paesmans M, Berghmans T, et al. Role of p53 as a prognostic factor for survival in lung cancer: a systematic review of the literature with a meta-analysis. Eur Respir J. 2001;18:705–719. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. | ||
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. | ||
DerSimonian R. Meta-analysis in the design and monitoring of clinical trials. Stat Med. 1996;15:1237–1248; discussion 49–52. | ||
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. | ||
Haroun RA, Zakhary NI, Mohamed MR, Abdelrahman AM, Kandil EI, Shalaby KA. Assessment of the prognostic value of methylation status and expression levels of FHIT, GSTP1 and p16 in non-small cell lung cancer in Egyptian patients. Asian Pac J Cancer Prev. 2014;15:4281–4287. | ||
Li W, Deng J, Tang JX. Combined effects methylation of FHIT, RASSF1A and RARbeta genes on non-small cell lung cancer in the Chinese population. Asian Pac J Cancer Prev. 2014;15:5233–5237. | ||
Zochbauer-Muller S, Fong KM, Maitra A, et al. 5′ CpG island methylation of the FHIT gene is correlated with loss of gene expression in lung and breast cancer. Cancer Res. 2001;61:3581–3585. | ||
Hsu HS, Chen TP, Hung CH, et al. Characterization of a multiple epigenetic marker panel for lung cancer detection and risk assessment in plasma. Cancer. 2007;110:2019–2026. | ||
Yanagawa N, Tamura G, Oizumi H, Endoh M, Sadahiro M, Motoyama T. Inverse correlation between EGFR mutation and FHIT, RASSF1A and RUNX3 methylation in lung adenocarcinoma: relation with smoking status. Anticancer Res. 2011;31:1211–1214. | ||
Song H, Yi J, Zhang Y, Wang R, Chen L. [DNA methylation of tumor suppressor genes located on chromosome 3p in non-small cell lung cancer]. Zhongguo Fei Ai Za Zhi. 2011;14:233–238. Chinese. | ||
Li W, Deng J, Jiang P, Tang J. Association of 5′-CpG island hypermethylation of the FHIT gene with lung cancer in southern-central Chinese population. Cancer Biol Ther. 2010;10:997–1000. | ||
Li H, Zhang W, Li W, Yin C. [Effects of methylation of FHIT gene on it’s protein and mrna expression in non-small cell lung cancer]. Zhongguo Fei Ai Za Zhi. 2009;12:760–764. Chinese. | ||
Yanagawa N, Tamura G, Oizumi H, et al. Promoter hypermethylation of RASSF1A and RUNX3 genes as an independent prognostic prediction marker in surgically resected non-small cell lung cancers. Lung Cancer. 2007;58:131–138. | ||
Kim DS, Cha SI, Lee JH, et al. Aberrant DNA methylation profiles of non-small cell lung cancers in a Korean population. Lung Cancer. 2007;58:1–6. | ||
Kim JS, Kim JW, Han J, Shim YM, Park J, Kim DH. Cohypermethylation of p16 and FHIT promoters as a prognostic factor of recurrence in surgically resected stage I non-small cell lung cancer. Cancer Res. 2006;66:4049–4054. | ||
Nakata S, Sugio K, Uramoto H, et al. The methylation status and protein expression of CDH1, p16(INK4A), and fragile histidine triad in nonsmall cell lung carcinoma: epigenetic silencing, clinical features, and prognostic significance. Cancer. 2006;106:2190–2199. | ||
Iliopoulos D, Guler G, Han SY, et al. Fragile genes as biomarkers: epigenetic control of WWOX and FHIT in lung, breast and bladder cancer. Oncogene. 2005;24:1625–1633. | ||
Tomizawa Y, Iijima H, Nomoto T, et al. Clinicopathological significance of aberrant methylation of RARbeta2 at 3p24, RASSF1A at 3p21.3, and FHIT at 3p14.2 in patients with non-small cell lung cancer. Lung Cancer. 2004;46:305–312. | ||
Tzao C, Tsai HY, Chen JT, Chen CY, Wang YC. 5′CpG island hypermethylation and aberrant transcript splicing both contribute to the inactivation of the FHIT gene in resected non-small cell lung cancer. Eur J Cancer. 2004;40:2175–2183. | ||
Kim JS, Kim H, Shim YM, Han J, Park J, Kim DH. Aberrant methylation of the FHIT gene in chronic smokers with early stage squamous cell carcinoma of the lung. Carcinogenesis. 2004;25:2165–2171. | ||
Maruyama R, Sugio K, Yoshino I, Maehara Y, Gazdar AF. Hypermethylation of FHIT as a prognostic marker in nonsmall cell lung carcinoma. Cancer. 2004;100:1472–1477. | ||
Jeong YJ, Jeong HY, Lee SM, Bong JG, Park SH, Oh HK. Promoter methylation status of the FHIT gene and Fhit expression: association with HER2/neu status in breast cancer patients. Oncol Rep. 2013;30:2270–2278. | ||
Al-Temaimi RA, Jacob S, Al-Ali W, Thomas DA, Al-Mulla F. Reduced FHIT expression is associated with mismatch repair deficient and high CpG island methylator phenotype colorectal cancer. J Histochem Cytochem. 2013;61:627–638. | ||
Banzai C, Nishino K, Quan J, et al. Promoter methylation of DAPK1, FHIT, MGMT, and CDKN2A genes in cervical carcinoma. Int J Clin Oncol. 2014;19:127–132. | ||
Paluszczak J, Misiak P, Wierzbicka M, Wozniak A, Baer-Dubowska W. Frequent hypermethylation of DAPK, RARbeta, MGMT, RASSF1A and FHIT in laryngeal squamous cell carcinomas and adjacent normal mucosa. Oral Oncol. 2011;47:104–107. | ||
Yin DT, Wang L, Sun J, et al. Association of the promoter methylation and protein expression of Fragile Histidine Triad (FHIT) gene with the progression of differentiated thyroid carcinoma. Int J Clin Exp Pathol. 2010;3:482–491. | ||
Yanagawa N, Osakabe M, Hayashi M, Tamura G, Motoyama T. Frequent epigenetic silencing of the FHIT gene in penile squamous cell carcinomas. Virchows Arch. 2008;452:377–382. | ||
Lee EJ, Lee BB, Kim JW, et al. Aberrant methylation of Fragile Histidine Triad gene is associated with poor prognosis in early stage esophageal squamous cell carcinoma. Eur J Cancer. 2006;42:972–980. | ||
Zheng S, Ma X, Zhang L, et al. Hypermethylation of the 5′ CpG island of the FHIT gene is associated with hyperdiploid and translocation-negative subtypes of pediatric leukemia. Cancer Res. 2004;64:2000–2006. | ||
Cecener G, Tunca B, Egeli U, et al. The promoter hypermethylation status of GATA6, MGMT, and FHIT in glioblastoma. Cell Mol Neurobiol. 2012;32:237–244. | ||
Wali A. FHIT: doubts are clear now. ScientificWorldJournal. 2010;10:1142–1151. | ||
Andriani F, Roz E, Caserini R, et al. Inactivation of both FHIT and p53 cooperate in deregulating proliferation-related pathways in lung cancer. J Thorac Oncol. 2012;7:631–642. | ||
Takada S, Morita K, Hayashi K, et al. Methylation status of fragile histidine triad (FHIT) gene and its clinical impact on prognosis of patients with multiple myeloma. Eur J Haematol. 2005;75:505–510. | ||
Wu Q, Shi H, Suo Z, Nesland JM. 5′-CpG island methylation of the FHIT gene is associated with reduced protein expression and higher clinical stage in cervical carcinomas. Ultrastruct Pathol. 2003;27:417–422. | ||
Shimada Y, Sato F, Watanabe G, et al. Loss of fragile histidine triad gene expression is associated with progression of esophageal squamous cell carcinoma, but not with the patient’s prognosis and smoking history. Cancer. 2000;89:5–11. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.









