Back to Journals » Journal of Blood Medicine » Volume 11
Serum Expression of Seven MicroRNAs in Chronic Lymphocytic Leukemia Patients
Authors Farzadfard E, Kalantari T, Tamaddon G
Received 12 September 2019
Accepted for publication 4 February 2020
Published 12 March 2020 Volume 2020:11 Pages 97—102
DOI https://doi.org/10.2147/JBM.S230842
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Ehsan Farzadfard,1 Tahereh Kalantari,2 Gholamhossein Tamaddon3,4
1School of Paramedical Sciences, Shiraz University of Medical Sciences, Shiraz, Iran; 2Department of Medical Biotechnology, School of Paramedical Sciences, Shiraz University of Medical Sciences, Shiraz, Iran; 3Department of Clinical Laboratory Sciences, School of Paramedical Sciences, Shiraz University of Medical Sciences, Shiraz, Iran; 4Diagnostic Laboratory Sciences and Technology Research Center, School of Paramedical Sciences, Shiraz University of Medical Sciences, Shiraz, Iran
Correspondence: Gholamhossein Tamaddon Building 99, Narvan Street, Shiraz 74616-86688, Tel +98 71 3227 2315
Fax +98 71 3227-0238
Email [email protected]
Purpose: MicroRNAs are small single-strand noncoding RNAs that can be deregulated in a variety of cancers. Over the past few years, multiple markers have been discovered in chronic lymphocytic leukemia (CLL). Among these, miRNAs seem to have important roles in the pathogenesis of CLL. The development and validation of miRNA-expression patterns as biomarkers should have a significant impact in cancer diagnosis, therapeutic success, and increasing the life expectancy of patients. In this study, to specify the utility of circulatory miRNA expression as noninvasive and useful biomarkers for CLL, we analyzed the dysregulation of seven miRNAs: miR30d, miR25-3p, miR19a-3p, miR133b, miR451a, miR145, and miR144 in CLL-patient sera.
Methods: Thirty untreated patients with flow-cytometry confirmation of CLL were chosen. Serum samples were collected from 30 newly diagnosed CLL patients. Fifteen healthy samples were taken for comparison as controls. RNA was extracted using Trizol. RNA from CLL patient specimens was compared to controls with real-time PCR.
Results: Seven miRNAs were differently expressed between CLL and normal specimens using the comparative 2−ΔΔCt method. miRNAs 133b, 25-3p, 451a, 145, 19a-3p, and 144 were overexpressed in sera obtained from CLL patients, and miRNA-30d was underexpressed in patient samples. Among these seven miRNAs, miR19a-3p and miR25-3p showed the most deregulation in CLL patients.
Conclusion: Real-time PCR is an applied means to perform high-throughput investigation of serum-RNA samples. We assessed the expression of seven miRNAs in CLL patients by this method. The results demonstrated that the use of miRNA-expression profiling may have an impressive role in the diagnosis of CLL. In addition, miRNA 19a-3p and 25-3p are known oncogenes with therapeutic and potential biomarkers.
Keywords: chronic lymphocytic leukemia, circulatory microRNAs, noncoding RNA, real-time PCR
Introduction
B-cell chronic lymphocytic leukemia (CLL) is the most common leukemia in adults, and is characterized by the expansion of CD5+/CD19+/CD23+ B lymphocytes.1,2 CLL has a heterogeneous clinical course in which patients show an accumulation of a malignant clone of B lymphocytes in their bone marrow, lymphatic tissue, or bloodstream.3 Several prognostic factors predicting the clinical course have been defined for CLL patients. There are some poor prognostic factors, such as mutations in IVGH, high-level expression of ZAP70 and surface protein CD38, and chromosomal aberration indicating a poor prognosis. On the other hand, patients with mutated IGVH or those lacking ZAP70 have a good prognosis.4–6
Clinical signs at the time of diagnosis are widely variable, and present as an indolent or aggressive state with different output. A large percentage of CLL patients are asymptomatic, and there is no need for treatment. Tiredness, weakness, anemia, lymphadenopathies, and splenomegaly are the main clinical signs in symptomatic patients who should receive treatment. Blood-cell counts, morphology of peripheral blood smears, and immunophenotyping are used for early diagnosis of CLL patients, but for further diagnosis more evaluation is required, such as expression of ZAP70 and CD38, which correlate with IGVH-mutation status.7,8 Also, cytogenetic and fluorescent in situ hybridization (FISH) are performed for CLL subtypes.9 However, the association between expression of ZAP70 or CD38 with unmutated IGVH genes is not absolute.10,11 On the other hand, FISH and bone-marrow biopsy, the key tests to choose appropriate treatment for patients with CLL, are invasive and expensive procedures. Therefore, it seems that the search for new biomarkers for CLL is necessary.12
Apoptosis is a physiological cell-suicide program that is one of the hallmarks of CLL, and impaired apoptosis represents an important mechanism in clinical resistance to therapies.13 Therefore, the development of therapeutic strategies based on targeting apoptosis in CLL is a very important issue. One of the important biomolecules that regulate the apoptosis pathway are miRNAs. MicroRNAs are a class of conserved small (~22 nt) noncoding RNAs that regulate gene expression at a posttranscriptional level by blocking translation or degrading target mRNAs. They can regulate many target genes simultaneously.14 Human miRNAs are located in cancer-associated regions of the genome. It seems that miRNAs play an important role in the pathogenesis of various human cancers.15–17 Evidence showed miRNAs can act as a diagnosis and therapeutic biomarker.18 Recently CD49d expression reported as a marker to decide for first treatment still, yet miRNAs be able to tools for biological treatment in addition biomarker.19, 20
The aim of this study was to investigate the expression of a group of miRNAs in CLL patients. Seven miRNAs — miR30d, miR25-3p, miR19a-3p, miR133b, miR451a, miR145, and miR144 — that target important genes in the apoptosis pathway and are deregulated in several hematologic disorderswere chosen for this study to consider their possible role as biomarkers. In the current study, we report the expression levels of these miRNAs detected by RT-PCR in serum samples of CLL patients with existing clinical data.
Methods
Samples and Patients
In accordance with the approval and moral-satisfaction form from Shiraz University of Medical Sciences (IR.SUMS.REC.1395.S609), 30 newly diagnosed CLL patients with >5,000/µL B lymphocytes in peripheral blood previously confirmed by flow cytometry between2016 and 2017 were enrolled in this study. Inclusion criteria were clinical signs and diagnosis of CLL and first diagnosis with no previous treatment. Serum samples from 15 healthy volunteers were used as controls. The control group underwent medical examinations and did not show any hematologic or other cancerous diseases.
RNA Extraction and cDNA Synthesis
Total RNA was extracted from the serum samples using Trizol reagent according to the manufacturer's protocol (Invitrogen, Carlsbad, CA, USA). Concentrations of extracted RNA were determined using NanoDrop (Hellma, Denmark), and aliquots of the samples were stored at −80°C. Specific cDNAs of selected miRNA were synthesized using specific primers (Parsgenome, Tehran, Iran) and a cDNA-synthesis kit (Exiqon, Vedbaek, Denmark). First, polyA tail was added to miRNAs with polyA polymerase at 37°C. Following this, RT enzyme, reaction buffer, and miR-specific primers for cDNA synthesis were mixed with RNA polyA tail, incubated at 45°C for 60 minutes, and inactivated at 85°C for 1 minute.
Real-Time PCR
Aliquots of the cDNA were used for quantitative PCR with real-time PCR Master Mix (Exiqon using an ABI (Applied Biosystems) apparatus according to the manufacturer’s instructions. Primer pairs were obtained commercially from Parsgenome). 5s rRNA was used as internal control and PCR normalization. qRT-PCR was run under conditions of initial denaturation at 95°C for 5 minutes, followed by 40 cycles of 95°C for 5 seconds, 63°C for 20 seconds, and 72°C for 30 seconds. All qRT-PCR tests were performed in duplicate.
Statistical Analysis
miRNA expression was calculated using the equation 2−ΔΔCt. Statistical analysis was performed by GraphPad Prism 5 (GraphPad Software, San Diego, CA, USA). miRNA expressions were compared between patient and control groups with t-tests. P<0.05 was considered statistically significant.
Bioinformatic Analysis
Identification of putative and validated target genes among differentially expressed genes for all seven miRNAs studied was performed using web-based software. The corresponding gene, miRBase ID, and sequence of each miRNA in this study was assigned before analysis. The web-based software used to investigate the miRNA targets was miRTarBase (http://mirtarbase.mbc.nctu.edu.tw), miRecords (http://c1.accurascience.com/miRecords), TargetScan (http://www.targetscan.org), miRanda (http://www.miRNA.org), DIANA microT (http://diana.imis.athena-innovation.gr/DianaTools), and miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2).
Results
Analysis of Serum-miRNA Expression in CLL Patients and Healthy Controls
To considering circulating miRNAs as potential diagnostic markers, we evaluatedcorrelations between the expression levels of seven circulating miRNAs (miR30d, miR25-3p, miR19a-3p, miR133b, miR451a, miR145, and miR144a) and of CLL diagnosis by comparison of CLL patients and healthy controls (Figure 1). To evaluate expression patterns of these seven miRNAs, we performed real-time PCR using the sera of 30 CLL patients and 15 healthy controls. Real-time PCR showed overexpression in miRNAs 25-3p, 19a-3p, 145, and 144 (P=0.0001, P=0.0007, P=0.023, P=0.0244) and lower expression of miRNAs-133b, 30d, and 451a (P=0.044, P=0.038, P=0.0323) in the sera of CLL patients compared to controls were found. Fold changes in median expression of the seven miRNAs in patient samples versus controls are given in Figure 2.
 |
Figure 1 Differentially expressed miRNAs in CLL cells and normal cells. |
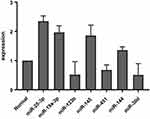 |
Figure 2 Median fold–change differences in alteration of miRNA expression between patient and control (normal). microRNAmiRNA expression analyzed using 2−ΔΔCt. |
Circulating miR25-3p Significantly Overexpressed in CLL Patients
Comparison of expression levels of the miR25-3p in sera of CLL patients and the control group is shown in Figure 3. As expected, real time PCR results showed significant alteration in miR25-3p expression in CLL samples. This result indicated that miR25-3p expression was much higher in CLL patients than healthy individuals — 2.35-fold (P=0.0001).
Circulating miR19a-3p Expression Higher in CLL Patients Than Healthy Controls
miR19a-3p (a member of the 17–92 miRNA cluster) expression levels, were demonstrated to be overexpressed in CLL patients compared with the control group. miR19a-3p expression was significantly upregulated in the sera of CLL patients compared with healthy individuals — 1.965-fold (P=0.0007, Figure 3).
Discussion
Discovering miRNAs and being more aware of their functions has accelerated our understanding about the mechanisms of regulation of gene expression. By binding to the 3ˊ UTR of their targets, miRNAs can posttranscriptionally regulate gene expression.21 Some evidence has shown that miRNAs play important roles in a variety of vital cell processes, such as growth, development, proliferation, and death,22,23 as well as pathological processes, including developmental abnormalities, cardiovascular and autoimmune diseases, and cancers.24,25
In this study, we showed different expressions of the seven miRNAs between CLL and normal seray using real-time PCR (Figure 1). Therefore, it seems that miRNA expression in serum samples can be turned into a noninvasive state to help diagnosis and treatment of CLL. We performed a pilot study to demonstrate that these miRNAs have a potential application as biomarkers for further studies in noninvasive CLL diagnosis and prospective studies with therapeutic targets.
As previously shown by other reports, many miRNAs are aberrantly expressed in CLL patients.26–28 In this study, we created an miRNA-expression profile that was achieved by investigating a set of 30 patients and 15 healthy controls. Cytogenetic disorders in CLL patients show that the deficiency in apoptotic processes usually happens in this leukemia. Bioinformatic analysis of our seven selected miRNAs revealed that these miRNAs have multiple powerful targets in the apoptosis pathway (Table 1). Among these miRNAs, two of four overexpressed miRNAs (miR19a-3p and miR25-3p) are potential oncomiRs whose functions as an oncomiR have been reported in a variety of cancers.29,30 In addition, they have great potential as diagnostic and therapeutic tools for CLL patients.
 |
Table 1 Intracellular Targets and Functions of miRNAs |
Overexpression of miR25-3p has been demonstrated in a variety of cancers, such as papillary thyroid carcinoma,31 osteosarcoma,32 hepatocellular carcinoma,32 and lung cancer.33 Heterogeneity of CLL has an impact on miR25-3p deregulation, as the expression level of miR25-3p has a correlation with such prognostic factors as cytogenetic disorders (lower levels in trisomy 12), RB1-gene deletion, and the age of patients.34 Our results showed the expression of this miRNA increased 2.35-fold (P=0.0001) in patient samples. This alteration shows the high-impact roles of miR25-3p as an oncomiR in the pathogenesis of CLL. TargetScan, miRanda, and miRTarBase analysis showed that it can potentially modulate the expression of important genes involved in regulating apoptosis and cell death (Table 1). All in all, this miRNA can be a suitable target to become a diagnostic biomarker or therapeutic tool in future studies.
miR19a-3p is a part of the 17–92 cluster, known as the oncomiR cluster.35,36 Overexpression of this miRNA cluster has been reported in several malignancies, such as acute myeloid leukemia,37 multiple myeloma,38 gastric cancer, and lung cancer.39,40 Bioinformatic analysis revealed that miR19a-3p has important putative targets, including PTEN, BIM, and TGFBR, in invasion, apoptosis, and cell death. In this study, high expression of miR19a-3p in CLL samples (1.965-fold, P=0.0007) shows the oncomiRic roles of miR19a-3p in CLL malignancy. The important targets in the apoptosis pathway indicate that this miRNA can play a significant role in pathogenesis and progression of CLL.
Both miR144 and -145 were overexpressed in CLL samples (1.36- and 1.86-fold, P=0.024, P=0.023), which is in accordance with their putative and validated targets, such as PTEN, NOTCH1, CDKN1, and MYC. We showed low expression of miR30d, 133b, and 451a (0.51-, 0.52-, and 0.68fold; P=0.038, P=0.044, P=0.032) was tumor-suppressive in the patient group, as previously demonstrated by other reports.41–43 Our data show that circulating miRNAs might have great potential as diagnostic and therapeutic biomarkers in CLL. Although our results suggest the mentioned roles for miR25-3p and miR19a-3p, further investigations are needed.
Conclusion
In the present study, we used real-time PCR to assess the expression of seven circulating miRNAs — miRNAs 30d, 25-3p, 19a-3p, 133b, 451a, 145, and 144 — in the sera of CLL patients. The results showed these miRNAs were dysregulated in CLL samples compared with healthy controls. We also showed that miR19a-3p and miR25-3p were significantly increased in CLL patients and that they can play important roles in the pathogenicity of CLL.
Acknowledgments
The authors thank all the participants in this study, as well as the nursing staff of the Leukemia Ward at Namazi Hospital. This article is part of Ehsan Farzadfard’s thesis for an MSc in Medical Biotechnology using a Shiraz University of Medical Sciences grant (95-01-10-11314).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Redaelli A, Laskin BL, Stephens JM, Botteman MF, Pashos CL. The clinical and epidemiological burden of chronic lymphocytic leukaemia. Eur J Cancer Care (Engl). 2004;13(3):279–287. doi:10.1111/ecc.2004.13.issue-3
2. Damle RN, Ghiotto F, Valetto A, et al. B-cell chronic lymphocytic leukemia cells express a surface membrane phenotype of activated, antigen-experienced B lymphocytes: presented in part at the 42nd annual meeting of the American Society of Hematology, December 1- 5, 2000, San Francisco, CA. Blood. 2002;99(11):4087–4093. doi:10.1182/blood.V99.11.4087
3. Zenz T, Mertens D, Küppers R, Döhner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer. 2010;10(1):37. doi:10.1038/nrc2764
4. Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia: presented in part at the 40th Annual Meeting of The American Society of Hematology, held in Miami Beach, FL, December 4- 8, 1998. Blood. 1999;94(6):1840–1847. doi:10.1182/blood.V94.6.1840
5. Wiestner A, Rosenwald A, Barry TS, et al. ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile. Blood. 2003;101(12):4944–4951. doi:10.1182/blood-2002-10-3306
6. Juliusson G, Oscier DG, Fitchett M, et al. Prognostic subgroups in B-cell chronic lymphocytic leukemia defined by specific chromosomal abnormalities. N Engl J Med. 1990;323(11):720–724. doi:10.1056/NEJM199009133231105
7. Abbott BL. Chronic lymphocytic leukemia: recent advances in diagnosis and treatment. Oncologist. 2006;11(1):21–30. doi:10.1634/theoncologist.11-1-21
8. Kröber A, Seiler T, Benner A, et al. V H mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100(4):1410–1416. doi:10.1182/blood.V100.4.1410.h81602001410_1410_1416
9. Mayr C, Speicher MR, Kofler DM, et al. Chromosomal translocations are associated with poor prognosis in chronic lymphocytic leukemia. Blood. 2006;107(2):742–751. doi:10.1182/blood-2005-05-2093
10. Grever MR, Lucas DM, Dewald GW, et al. Comprehensive assessment of genetic and molecular features predicting outcome in patients with chronic lymphocytic leukemia: results from the US intergroup Phase III trial E2997. J Clin Oncol. 2007;25(7):799–804. doi:10.1200/JCO.2006.08.3089
11. Byrd JC, Gribben JG, Peterson BL, et al. Select high-risk genetic features predict earlier progression following chemo immunotherapy with fludarabine and rituximab in chronic lymphocytic leukemia: justification for risk-adapted therapy. J Clin Oncol. 2006;24(3):437–443. doi:10.1200/JCO.2005.03.1021
12. Alsagaby SA, Brennan P, Pepper C. Key molecular drivers of chronic lymphocytic leukemia. Clin Lymphoma Myeloma Leuk. 2016;16(11):593–606. doi:10.1016/j.clml.2016.08.008
13. Billard C. Apoptosis inducers in chronic lymphocytic leukemia. Oncotarget. 2014;5(2):309. doi:10.18632/oncotarget.v5i2
14. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
15. McManus MT. MicroRNAs and cancer. Semin Cancer Biol. 2003;13(4):253–258. doi:10.1016/S1044-579X(03)00038-5
16. Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi:10.1146/annurev.med.59.053006.104707
17. Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi:10.1073/pnas.0307323101
18. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi:10.1016/j.cell.2009.01.002
19. Baumann T, Delgado J, Santacruz R, et al. CD 49d (ITGA 4) expression is a predictor of time to first treatment in patients with chronic lymphocytic leukaemia and mutated IGHV status. Br J Haematol. 2016;172(1):48–55. doi:10.1111/bjh.13788
20. Alsagaby SA, Alhumaydhi FA. Proteomics insights into the pathology and prognosis of chronic lymphocytic leukemia. Saudi Med J. 2019;40(4):317–327. doi:10.15537/smj.2019.4.23598
21. Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310(2):442–453. doi:10.1016/j.ydbio.2007.08.007
22. He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130. doi:10.1038/nature05939
23. Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469(7330):336. doi:10.1038/nature09783
24. Nakasa T, Miyaki S, Okubo A, et al. Expression of microRNA‐146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008;58(5):1284–1292. doi:10.1002/art.23429
25. Sandhu SK, Croce CM, Garzon R. Micro-RNA expression and function in lymphomas. Adv Hematol. 2011;2011:1–12. doi:10.1155/2011/347137
26. Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24):15524–15529. doi:10.1073/pnas.242606799
27. Visone R, Veronese A, Balatti V, Croce CM. MiR-181b: new perspective to evaluate disease progression in chronic lymphocytic leukemia. Oncotarget. 2012;3(2):195.
28. Balatti V, Pekarky Y, Croce CM. Role of microRNA in chronic lymphocytic leukemia onset and progression. J Hematol Oncol. 2015;8(1):12. doi:10.1186/s13045-015-0112-x
29. Feng Y, Liu J, Kang Y, et al. miR-19a acts as an oncogenic microRNA and is up-regulated in bladder cancer. J Exp Clin Cancer Res. 2014;33(1):67. doi:10.1186/s13046-014-0067-8
30. Fujiwara T, Uotani K, Yoshida A, et al. Clinical significance of circulating miR-25-3p as a novel diagnostic and prognostic biomarker in osteosarcoma. Oncotarget. 2017;8(20):33375. doi:10.18632/oncotarget.v8i20
31. Li M, Song Q, Li H, Lou Y, Wang L, Ray RB. Circulating miR-25-3p and miR-451a may be potential biomarkers for the diagnosis of papillary thyroid carcinoma. PLoS One. 2015;10(7):e0132403. doi:10.1371/journal.pone.0132403
32. Jin Y, Yu D, Tolleson WH, et al. MicroRNA hsa-miR-25-3p suppresses the expression and drug induction of CYP2B6 in human hepatocytes. Biochem Pharmacol. 2016;113:88–96. doi:10.1016/j.bcp.2016.06.007
33. Xiang J, Hang JB, Che JM, Li HC. miR-25 is up-regulated in non-small cell lung cancer and promotes cell proliferation and motility by targeting FBXW7. Int J Clin Exp Pathol. 2015;8(8):9147.
34. Filip AA, Grenda A, Popek S, et al. Expression of circulating miRNAs associated with lymphocyte differentiation and activation in CLL—another piece in the puzzle. Ann Hematol. 2017;96(1):33–50. doi:10.1007/s00277-016-2840-6
35. He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828. doi:10.1038/nature03552
36. Olive V, Bennett MJ, Walker JC, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23(24):2839–2849. doi:10.1101/gad.1861409
37. Li Z, Lu J, Sun M, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci USA. 2008;105(40):15535–15540. doi:10.1073/pnas.0808266105
38. Chen L, Li C, Zhang R, et al. miR-17-92 cluster microRNAs confers tumorigenicity in multiple myeloma. Cancer Lett. 2011;309(1):62–70. doi:10.1016/j.canlet.2011.05.017
39. Li H, Wu Q, Li T, et al. The miR-17-92 cluster as a potential biomarker for the early diagnosis of gastric cancer: evidence and literature review. Oncotarget. 2017;8(28):45060.
40. Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65(21):9628–9632. doi:10.1158/0008-5472.CAN-05-2352
41. Zhang R, Xu J, Zhao J, Bai J. Mir-30d suppresses cell proliferation of colon cancer cells by inhibiting cell autophagy and promoting cell apoptosis. Tumour Biol. 2017;39(6):1010428317703984. doi:10.1177/1010428317703984
42. Xiang KM, Li XR. MiR-133b acts as a tumor suppressor and negatively regulates TBPL1 in colorectal cancer cells. Asian Pac J Cancer Prev. 2014;15(8):3767–3772. doi:10.7314/APJCP.2014.15.8.3767
43. Li X, Sanda T, Look AT, Novina CD, von Boehmer H. Repression of tumor suppressor miR-451 is essential for NOTCH1-induced oncogenesis in T-ALL. J Exp Med. 2011;208(4):663–675. doi:10.1084/jem.20102384
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

