Back to Journals » International Journal of Nanomedicine » Volume 14
Selenium Nanoparticle Protection of Fibroblast Stress: Activation of ATF4 and Bcl-xL Expression
Authors Chung S, Roy AK, Webster TJ
Received 25 April 2018
Accepted for publication 30 August 2018
Published 20 December 2019 Volume 2019:14 Pages 9995—10007
DOI https://doi.org/10.2147/IJN.S172236
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Carlos Rinaldi
Stanley Chung, Amit K Roy, Thomas J Webster
Department of Chemical Engineering, Northeastern University, Boston, MA 02115, USA
Correspondence: Thomas J Webster
360 Huntington Avenue, 313 Snell Engineering Center, Boston, MA 02215, USA
Tel +1-617-373-6585
Email [email protected]
Background: In recent years, selenium nanostructures have been researched due to their antibacterial properties, low toxicity to mammalian cells, and high biological efficacy. However, the clinical implementation of the use of selenium has received mixed results, and there is much work needed to improve the understanding of the biological mechanisms involved in the observed cellular responses.
Materials and methods: In this work, an investigation into the mechanistic pathways of selenium nanoparticles (SeNPs) in biological systems was conducted by studying the changes in gene expression of ATF4, Bcl-xL, BAD2, HSP70, and SOD2 in non-cancerous human dermal fibroblasts (HDF) under oxidative stress, nutrient deprivation stress, and no treatment (control) conditions.
Results: This study revealed that SeNP incubation led to reduced internal reactive oxygen species (ROS) generation for all conditions tested, thus, providing a protective environment for HDF. At the stress conditions, the expression of ATF4 and Bcl-xL increased for cells treated with SeNP incubation, leading to attenuation of the cells under stress. These results also hint at reductive stress causing a detrimental impact to cell proliferation under routine cell passaging conditions.
Conclusion: In summary, this study highlights some possible mechanistic pathways implicated in the action of SeNPs that warrant further investigation (specifically, reducing stress conditions for HDF) and continues to support the promise of SeNPs in a wide range of medical applications.
Keywords: selenium nanoparticles, oxidative stress, reductive stress, anti-apoptosis, stress response
Introduction
Selenium (Se) materials have shown great promise for a variety of clinical applications most notably as a vitamin supplement but also for the management of oxidative stress1–4 and reduction of cancer5–9 and bacterial cells,10–14 and have shown improved efficacy and safety compared to larger sized Se compounds.1–7,15–17 Previous research has shown some selectivity towards inhibiting bacterial growth while maintaining or enhancing mammalian proliferation when exposed to selenium nanoparticles. Selenium is a common trace element in the body that is essential to healthy nutrition, due to the formation of selenoproteins18,19 and may even possess anticancer properties.6
The application of Se nanoparticles (SeNPs) is particularly promising as SeNPs possess lower toxicity than their elemental form; for example, in mice, the LD50 of selenomethionine was 25.6 mg Se/kg compared with 92.1 mg Se/kg for SeNPs.20 SeNPs also possess a number of material properties not present in the bulk form:21 SeNPs have a high surface area/volume ratio and possess increased surface area and increased interactions with biological targets, leading to higher bioavailability and absorption by the host.7,22,23 Researchers have shown that SeNPs may be used as a coating or directly in solution at dosages that reduce bacterial and cancerous growth while maintaining or promoting mammalian cell function.3,5,11–13,24
Despite their many advantages, SeNPs, like many other metallic nanoparticles, may induce oxidative stress and produce reactive oxygen species at high concentrations.25 Thus, Se chemistry presents an interesting paradox between both pro- and anti-oxidant activities, depending on the dosage and cell types exposed.26,27 Se compounds may cause oxidative stress, DNA damage, and apoptosis8,9,28–30 or rescue cells from oxidative damage,31,32 heat shock,1 cadmium toxicity,33,34 and neural toxins.35 In an attempt to clarify the mechanistic effect of SeNPs on biological systems, the study presented here investigated changes in gene expression of human dermal fibroblasts (HDF) during two different stressed conditions: oxidative stress and nutrient deprivation, with and without SeNP pre-incubation. The changes in the expression of ATF4, Bcl-xL, BAD2, HSP70, and SOD2 in fibroblasts were studied to assess the response to different stress conditions and to a no treatment control. Importantly, to the best of the authors’ knowledge, this represents the first molecular mechanism study of healthy fibroblasts when exposed to SeNPs (and such data are often missing from the entire nanoparticle field in general).
Materials and Methods
Selenium Nanoparticle Synthesis
Bovine serum albumin (BSA, Sigma Aldrich, Saint Louis, MO)-coated SeNPs were synthesized with sodium selenite and ascorbic acid. 10 mL of 50 mM ascorbic acid (Sigma Aldrich) was added dropwise to 1 mL of an aqueous solution containing 100 mM sodium selenite and 10 mg/mL BSA at an agitation speed of 600 rpm. Upon the addition of the ascorbic acid to the sodium selenite, the solution turned into light yellow and eventually dark red. The reaction proceeded for 30 mins before collecting the NPs by centrifugation (11,000 rpm at 15 mins). NPs were washed at least twice by dI H2O (dI H2O, Milli-Q system, EMD Millipore, Billerica, MA).
Mammalian Cell Activity Culture
Passage 3–12 human dermal fibroblasts (HDF, Lonza, Basel, Switzerland; non-cancerous cell line) were cultured using Dulbecco’s Modified Eagle Medium (DMEM, Sigma Aldrich) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT) and 1% penicillin/streptomycin (P/S, Sigma Aldrich) in a 37°C, humidified, 5% CO2/95% air environment. HDFs were cultured to ~90% confluence, rinsed with Dulbecco’s phosphate-buffered saline (dPBS, Sigma Aldrich), and detached from the tissue culture surface with 0.25% trypsin-EDTA (Sigma-Aldrich). HDF cells were then seeded into 96 well plates at 5000 cells/well. After 6 hrs, DMEM with and without SeNPs were separately added to the cells for 24 hrs. The media was then exchanged with the stress conditions, either 150 μM hydroquinone (Sigma Aldrich), DMEM with only 0.2% FBS, or DMEM with 10% FBS (negative control). Cells were then cultured for an additional 72 hrs in the media containing the stressors.
Mammalian Cell Characterization Assays
Downstream assays were conducted at the 6, 12, 24, 48, and 72 hrs time points for the live/dead stain, intracellular ROS assay, or RNA extraction. The MTS assay was conducted at 24, 48, and 72 hrs. For qPCR studies, the cells were grown in a 6-well plate at 150,000 cells/well (scaled based on the surface area).
For determining cell count, the MTS assay was used (Cell Titer 96 Aqueous One Solution Cell Proliferation Assay, G3581 Promega). The cell culture medium was aspirated before adding a 1:5 (media: MTS) solution for 2.5 hrs and before measuring the absorbance of each well by a SpectraMaxM3 plate-reader at 490 nm. Controls included wells with no cells, and a standard curve was constructed with known cell numbers.
A live/dead stain (calcein AM/ethidium homodimer-1, Invitrogen) was used to assess the health of the cells. The cells were centrifuged at 200g for 5 mins before the addition of the dye (final concentration: calcein AM = 1 μM/ethidium homodimer-1 = 2 μM). The cells were then imaged using the Zeiss Axio Observer Z1 inverted microscope.
Intracellular ROS was measured using a CM-H2DCFDA kit (ThermoFisher). The HDF cells were washed with dPBS before incubating with 100 μM CM-H2DCFDA for 1 hr. Afterwards, the dye solution was removed, and the cells were washed once more with dPBS before incubating in DMEM for 24 hrs to develop the signal. The fluorescent signal was read by a SpectraMax M3 at Ex/Em: 495 nm/520 nm.
For all procedures involving RNA extraction, reverse transcriptase (RT)-PCR, and qPCR, only RNAse-DNAse-free supplies (pipette tips, centrifuge tubes, water, etc.) were used. A Trizol (ThermoFisher) reagent was used to extract the RNA from the HDF cells. After the designated incubation time, the media was aspirated, and the cells were washed 1x with dPBS. 0.5 mL of Trizol was then added to each well and transferred into a centrifuge tube. Chloroform (Sigma) was added to the solution of Trizol at a 1/5 volume ratio of the Trizol (100 μL). The samples were then centrifuged at 12,000 g for 15 min at 4°C. The clear aqueous phase was then transferred to new centrifuged tubes, and 300 μL of isopropanol (Sigma) was added to precipitate the RNA. The samples were centrifuged once more at 12,000 g for 10 min at 4°C. The supernatant was removed, and the precipitated RNA was washed with 75% ethanol, diluted with pure ethanol (Sigma) and DEPC-treated water (ThermoFisher), at equal volumes to the Trizol (500 μL). The samples were centrifuged at 12,000 g for 2 min at 4°C. Afterwards, the ethanol was decanted away; the RNA precipitate was air-dried on ice until most of the ethanol evaporated. Afterwards, the RNA was resuspended in 20 μL of DEPC-treated water in preparation for the RT-PCR reaction.
Reagents from the RevertAid RT Reverse Transcription Kit (ThermoFisher) were used to perform the RT-PCR reaction. 2 μL of RNA was used for each reaction to a total volume of 20 μL, following the manufacturer’s instructions. The RNA samples were then incubated at 25°C for 5 mins, heated to 45°C for 60 mins, and terminated at 70°C for 5 mins. 100 μL of DEPC-treated water was then added to the cDNA product before proceeding to qPCR studies.
The qPCR studies performed here utilized the SYBR green dye (ABI Prism, KAPA). Each reaction contained 5 μL of cDNA, 1 μL of forward and reverse primers at10 μM, the reaction master mix, and DEPC-treated water, totaling a 20 μL reaction volume. 6 genes (GAPDH, ATF4, Bcl-xL, BAD, SOD2, HSP70) were tested using the following primers:
GAPDH F: AAC GGG AAG CTT GTC ATC AAT GGA AA
GAPDH R: GCA TCA GCA GAG GGG GCA GAG
ATF4 F: ATG ACC GAA ATG AGC TTC CTG
ATF4 R: CTG GAG AAC CCA TGA GGT TTG
Bcl-xL F: GAT CCC CAT GGC AGC AGT AAA GCA AG
Bcl-xL R: CCC CAT CCC GGA AGA GTT CAT TCA CT
BAD F: CGC TAC GGT GGG AGA GGA AGC
BAD R: GCT CAC TCG GCT CAA ACT CTG GGA
SOD2 F: GGA AGC CAT CAA ACG TGA CTT
SOD2 R: CCC GTT CCT TAT TGA AAC CAA GC
HSP70 F: ACT CTT GAG GCC TAC TAC AGC TCT CC
HSP70 R: TGG AGC CAT CAG ACT GAG GAG TGA
The reaction took place inside a QuantStudio 6 (ThermoFisher) with the following heating protocol: 3 min hold at 95°C and 40 cycles of 95°C for 3 sec and 60°C for 20 sec. A melt curve followed every reaction to assess whether the reaction produced single, specific products. GAPDH was used as the endogenous control, and the gene expression was referenced against the HDFs without SeNP incubation and fed only 10% FBS DMEM. The relative quantification (RQ) was calculated by 2−ΔΔCT.
Statistical Analysis
All experiments were conducted in triplicate. Analysis of variance (ANOVA) and Student’s t-tests were used to determine the significance in the changes in cellular activity.
Results
Cell Viability and Morphology After Stress
The HDF cells were pre-incubated with 10 μg/mL SeNP based on previous experiments investigating the effect of dosing on HDF viability. This concentration represented the highest concentration at which there was a negligible detrimental impact to HDF proliferation. While clinically it is unlikely that 10 μg/mL will reach the cells of interest, even an order magnitude of initial dosage larger (100 μg/mL) is within a reasonable dosage for Se in the body. The cells were challenged with either hydroquinone, a strong oxidizer, or a low nutrient condition (low serum) for D1, 2, and 3 after the 1 day incubation with SeNP (Figure 1).
Figure 1A shows that the addition of SeNP led to an approximate 20% reduction in MTS activity on all three days tested which was in line with previous results (p < 0.01). For the -SeNP condition (solid lines in Figure 1A–C), the D1 and D2 cell numbers (p > 0.05) for the normal and low serum conditions were almost identical until D3 (p < 0.01). This would suggest that the low serum did not fully affect the cells until D2. Interestingly, although pre-incubation with SeNP (dotted lines in Figure 1A–C) decreased the MTS signal on D1 and D2, the D3 signal was comparable to that of the -SeNP condition. Once nutrient depletion started to affect the cells, the +SeNP condition led to a lower drop in MTS activity. The HQ challenge caused a significant loss of MTS signal for both of the +SeNP and -SeNP conditions (Figure 1C). However, the +SeNP had approximately doubled the cell number compared to the -SeNP condition for all the time points tested (p < 0.01). Next, the dose response of the SeNP, ranging from 1–50 μg/mL, incubation was tested once more against the HQ challenge (Figure 1D). All the +SeNP conditions had a higher MTS response than the -SeNP condition although the MTS signal was inversely proportional to the concentration of SeNP used to incubate the cells. Interestingly, at the 20 and 50 μg/mL concentration of SeNPs, the cells lost the cytoprotective effect at 72 hrs and were not statistically different from that of no SeNP incubation (p > 0.05).
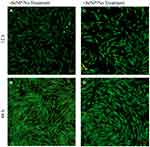
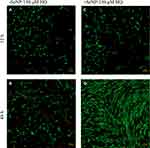
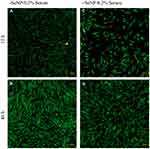
Both phase-contrast and live/dead stains of the cells were imaged to assess the cell morphology. For the no-treatment condition (Figures 2 and 3), the morphology of both the -SeNP and +SeNP conditions appeared normal and undamaged, exhibiting the expected triangular spreading from fibroblasts and high viability. The -SeNP conditions displayed irregular morphology: many cells were rounded (Figure 4A–D), and microscopy images from the live/dead stain indeed showed higher amounts of dead cells at the 48 hrs condition in the -SeNP condition (Figure 5A–D). Conversely, the live/dead images for the +SeNP condition showed generally a healthy morphology and high viability. Finally, the images for the low serum condition (Figures 6 and 7) showed no distinguishable difference.
ROS and Gene Expression
Intracellular ROS was measured using the CM-H2DCFDA kit. CM-H2DCFDA fluoresces upon cleavage by intracellular esterases, which have increased activity in oxidative stress conditions. In all +SeNP conditions, the amount of internal ROS was reduced, relative to the -SeNP condition, until 24 hrs, at which point the ROS became higher, relative to the -SeNP condition. The low serum condition and the no treatment conditions demonstrated comparable levels of ROS between both the -SeNP and +SeNP conditions until 72 hrs (Figure 8A and B). In the HQ challenge (Figure 8C), the +SeNP condition had approximately doubled the internal ROS by 72 hrs compared to the -SeNP condition.
qPCR studies were then conducted, measuring the relative quantification (RQ) of 5 different genes: ATF4, BAD, Bcl-xL, HSP70, and SOD2 (Figure 9). ATF4 had previously been shown to attenuate the cell response during hypoxia36 and had increased gene expression in a nutrient depletion model.37 The 12 and 48 hrs time points were chosen as those where the time points closest to the 24 hrs inflection where the +SeNP condition changed from a lower relative ROS to a higher relative ROS compared to the -SeNP condition. In both the no treatment and the HQ challenge condition, the addition of SeNP substantially elevated the activity of SOD2 at 12 hrs (Figure 9B) before falling back to baseline levels at 48 hrs (Figure 9E). In addition, in both of these conditions, the addition of SeNP led to a mildly elevated expression of HSP70 at 12 hrs (Figure 9B and E) before returning to the baseline. For the no-treatment condition, the addition of SeNP (Figure 9A) did not significantly affect the expression of ATF4, BAD, or Bcl-xL. At the stressed conditions (Figure 9B and C), the addition of SeNP led to a higher expression of ATF4 and Bcl-xL at 12 hrs. The expression of Bcl-xL remained elevated at 48 hrs (Figure 9E and F). BAD expression was elevated in both +SeNP/stressed conditions although not significantly above the -SeNP conditions. Surprisingly, HSP70 showed a lower expression in both +SeNP/stressed conditions at both 12 and 48 hrs time points.
Discussion
Based on the qPCR and internal ROS studies, the key findings for each of the stressor groups tested (no treatment, HQ, and 0.2% low serum) are summarized in Table 1 and are further explained below.
 |
Table 1 Summation of Significant Changes in Gene Expression of Cells with Selenium Nanoparticle (SeNP) Pre-Incubation Compared with Cells with No SeNP |
No Treatment Conditions
SeNP led to enhanced HSP70 and SOD2 gene expression at 12 hrs, perhaps a sign of the increased activation of the antioxidant pathway. By 48 hrs, the gene expression of both returned to their normal level (~1). The expression level of ATF4, BAD, and Bcl-xL at both time points did not conclusively differ from that of the -SeNP condition. However, based on the inflection in intracellular ROS behavior from relatively lower levels to relatively higher levels than the -SeNP conditions at around 24–48 hrs, it may be inferred that the increased SOD2 expression during the 12 hrs may have led to anti-oxidant depletion in the Se treated condition, causing the internal ROS to increase once the antioxidant stores were depleted. Studies involving other metallic nanoparticles have shown a similar phenomenon where activation in SOD and CAT have coincided with a depletion of GSH stores.25 In addition, excessive reducing equivalents, such as SOD2,38 may lead to reductive stress that harms cell culture performance. This may account for how higher doses of SeNP lead to poorer cell rescuing (Figure 1D) if the increased dosage overshot the oxidative balance towards reductive stress. Regardless, the homeostasis between pro-oxidant and antioxidant is a delicate balance where excessive signals from either direction may lead to detrimental stress for the cells.
HQ Condition
In response to HQ stress, SeNP activated the ATF4, SOD2, and Bcl-xL response to protect the cells at 12 hrs. Increased activation of ATF4 was expected as it was previously shown to respond to exogenous stresses.36,37 In this case, the activation of the anti-oxidative stress pathway was needed to counter the HQ stress and help increase cell numbers. Additionally, the intracellular ROS values for the +SeNP condition were reduced compared to the -Se condition until 24 hrs. However, this assay also measured only intracellular ROS as an absolute value, based on the number of viable cells. The +SeNP condition was approximately double the number of viable cells as the -Se condition at 48 and 72 hrs. Internal ROS values for the +Se condition were 120% for both 48 and 72 hrs and 84 and 74% for the -Se condition at 48 and 72 hrs, respectively. Together, this may indicate that the internal ROS per cell was likely even lower in the +SeNP condition and this may be why the cells were able to proliferate.
By 48 hrs, the internal antioxidant mechanism was likely depleted, leading to a reduced expression of SOD2. The expression of the BAD and Bcl-xL genes was at the same level on the Se treated cells from 12 to 48 hrs whereas the gene expression of these two genes increased on the untreated cells from 12 to 48 hrs. In the case of HQ stress, the likely mechanism to the rescuing effect was in the activation of the anti-oxidative stress pathways and the anti-apoptotic pathways.
0.2% Serum Condition
The 0.2% serum condition may be thought of as two phases: the first phase at D1 and 2 and the second phase at D3. In D1 and D2, the response was very similar to the response from the no-treatment condition: the cell numbers for the no treatment condition compared to the low serum condition were very similar at the D1 and D2 time points (Figure 1B), suggesting that nutrient depletion had yet to affect the cell proliferation. However, even though the cell growth was unaffected, the HDF cells already exhibited differential gene expression. Similar to the HQ challenge, the 0.2% serum condition increased expression of ATF4 and Bcl-xL for the +SeNP condition in response to this environmental shock.
Reduction in HSP70 and SOD expression was surprising as HSP70 may also regulate against nutrient depletion. In some cases though, researchers have observed no change to HSP70 after heat shock in the presence of Se.39 In that study, Rivera et al found that the turkey embryos compensated for the heat shock by increased GPx activity. GPx utilizes GSH as a substrate for converting H2O2 to water. GSH depletion was already suspected as the cause of the internal ROS inflection and may play a role in this current system. Hyperactive GSH would account for both the reduced HSP70 response and the reduced ROS levels in the +SeNP condition although further studies will be needed to confirm this theory.
By D3, the cell number of the -SeNP condition had fallen to the same level as that of the +SeNP condition, and the 48 hrs qPCR results indicated that both the -SeNP and +SeNP conditions had significant activation of the apoptotic pathways, as shown by the increase in BAD expression from 12 to 48 hrs. However, the +Se treated cells showed a lower activation of the BAD gene while maintaining a higher expression of the Bcl-xL gene, which may account for the reduced drop in cell number as compared with the -Se condition.
Conclusion
Results of the present study suggest that the biological action of SeNP may be caused primarily by changes in the oxidative state as well as activation of stress response and anti-apoptotic pathways. The ATF4 and Bcl-xL genes were both activated in the stress conditions in SeNP pre-incubated cells, and the attenuation in cell number in the +SeNP cells was statistically significant. The preliminary evidence here shows signs of reductive stress; this buildup of reducing equivalents may harm cells in the absence of external stressors but protects cells in the event of external stresses. There is a growing body of literature showing excessive reducing equivalents eventually lead to increased oxidative stress.40–42 It may be possible that the pro-oxidant effects induced by SeNPs shown in the literature were caused by the overactive reductive stress although further investigations will be needed to confirm this theory. Future studies would first focus on confirming the initial findings reported here.
Results from these studies hint at the presence of reductive stress, something previously only observed in cardiac systems. Implications of these results could change the paradigm on oxidative stress and all associated conditions, including aging, immunology, and cardiology, although further studies are needed to strengthen this claim. Additional studies would investigate the activity of the GSH/GPx system to confirm the presence of reductive stress. Assays looking at the ratio of the GSH (reduced form)/GSSG (oxidized form) and protein expression may provide further insight into the mechanism of SeNP activity.
Results from this work have also shown the sensitivity of cells to the dosing of SeNPs, and future studies should focus on determining the localization of the SeNPs in the cells. Information about the SeNP internalization would also help to address whether these changes in genetic activity were caused by antioxidant and stress pathway activation in response to SeNPs or by SeNP metabolism by the cells. Further studies investigating the protein species associated with Se metabolism would help isolate the contributions of the Se metabolism to the changes in gene expression shown here.
Acknowledgments
The authors would like to thank the Department of Chemical Engineering for funding and Mian Wang for his invaluable assistance and help.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Yuan B, Webster TJ, Roy AK. Cytoprotective effects of cerium and selenium nanoparticles on heat-shocked human dermal fibroblasts: an in vitro evaluation. Int J Nanomedicine. 2016;1427. doi: 10.2147/IJN.S104082.
2. Kalishwaralal K, Jeyabharathi S, Sundar K, Muthukumaran A. Sodium selenite/selenium nanoparticles (SeNPs) protect cardiomyoblasts and zebrafish embryos against ethanol induced oxidative stress. J Trace Elem Med Biol. 2015;32:135–144. doi:10.1016/j.jtemb.2015.06.010
3. Bhattacharjee A, Basu A, Sen T, Biswas J, Bhattacharya S. Nano-Se as a novel candidate in the management of oxidative stress related disorders and cancer. Nucl. 2017;60(2):137–145. doi:10.1007/s13237-016-0183-2
4. Shirsat S, Kadam A, Mane RS, et al. Protective role of biogenic selenium nanoparticles in immunological and oxidative stress generated by enrofloxacin in broiler chicken. Dalt Trans. 2016;45(21):8845–8853. doi:10.1039/c6dt00120c
5. Fernandes AP, Gandin V. Selenium compounds as therapeutic agents in cancer. Biochim Biophys Acta - Gen Subj. 2015;1850(8):1642–1660. doi:10.1016/j.bbagen.2014.10.008
6. Clark LC, Combs GF
7. Skalickova S, Milosavljevic V, Cihalova K, Horky P, Richtera L, Adam V. Selenium nanoparticles as a nutritional supplement. Nutrition. 2017;33:83–90. doi:10.1016/j.nut.2016.05.001
8. Wei Y, Cao X, Ou Y, Lu J, Xing C, Zheng R. SeO2 induces apoptosis with down-regulation of Bcl-2 and up-regulation of P53 expression in both immortal human hepatic cell line and hepatoma cell line. Mutat Res Genet Toxicol Environ Mutagen. 2001;490(2):113–121. doi:10.1016/S1383-5718(00)00149-2
9. Rudolf E, Rudolf K, Červinka M. Selenium activates p53 and p38 pathways and induces caspase-independent cell death in cervical cancer cells. Cell Biol Toxicol. 2008;24(2):123–141. doi:10.1007/s10565-007-9022-1
10. Shakibaie M, Forootanfar H, Golkari Y, Mohammadi-Khorsand T, Shakibaie MR. Anti-biofilm activity of biogenic selenium nanoparticles and selenium dioxide against clinical isolates of Staphylococcus aureus, Pseudomonas aeruginosa, and Proteus mirabilis. J Trace Elem Med Biol. 2015;29:235–241. doi:10.1016/j.jtemb.2014.07.020
11. Tran PA, Webster TJ. Selenium nanoparticles inhibit Staphylococcus aureus growth. Int J Nanomedicine. 2011;6:1553. doi:10.2147/IJN.S25646
12. Tran PA, Webster TJ. Antimicrobial selenium nanoparticle coatings on polymeric medical devices. Nanotechnology. 2013;24(15):155101. doi:10.1088/0957-4484/24/15/155101
13. Wang Q, Webster TJ. Short communication: inhibiting biofilm formation on paper towels through the use of selenium nanoparticles coatings. Int J Nanomedicine. 2013;8:407. doi:10.2147/IJN.S37465
14. Wang Q, Webster TJ. Nanostructured selenium for preventing biofilm formation on polycarbonate medical devices. J Biomed Mater Res Part A. 2012;100A(12):3205–3210. doi:10.1002/jbm.a.34262
15. Liu W, Golshan NH, Deng X, et al. Selenium nanoparticles incorporated into titania nanotubes inhibit bacterial growth and macrophage proliferation. Nanoscale. 2016;8(34):15783–15794. doi:10.1039/C6NR04461A
16. Sarkar B, Bhattacharjee S, Daware A, Tribedi P, Krishnani KK, Minhas PS. Selenium nanoparticles for stress-resilient fish and livestock. Nanoscale Res Lett. 2015;10(1):371. doi:10.1186/s11671-015-1073-2
17. Chaudhary S, Umar A, Mehta SK. Selenium nanomaterials: an overview of recent developments in synthesis, properties and potential applications. Prog Mater Sci. 2016;83:270–329. doi:10.1016/j.pmatsci.2016.07.001
18. Andrews PJD, Avenell A, Noble DW, et al. Randomised trial of glutamine, selenium, or both, to supplement parenteral nutrition for critically ill patients. Br Med J. 2011;342:d1542–d1542. doi:10.1136/Bmj.D1542
19. Santhosh Kumar B, Priyadarsini KI. Selenium nutrition: how important is it? Biomed Prev Nutr. 2014;4(2):333–341. doi:10.1016/j.bionut.2014.01.006
20. Wang H, Zhang J, Yu H. Elemental selenium at nano size possesses lower toxicity without compromising the fundamental effect on selenoenzymes: comparison with selenomethionine in mice. Free Radic Biol Med. 2007;42(10):1524–1533. doi:10.1016/j.freeradbiomed.2007.02.013
21. Whitesides GM. Nanoscience, nanotechnology, and chemistry. Small. 2005;1(2):172–179. doi:10.1002/smll.200400130
22. Chen G, Roy I, Yang C, Prasad PN. Nanochemistry and nanomedicine for nanoparticle-based diagnostics and therapy. Chem Rev. 2016;116(5):2826–2885. doi:10.1021/acs.chemrev.5b00148
23. Goldberg M, Langer R, Jia X. Nanostructured materials for applications in drug delivery and tissue engineering. J Biomater Sci Polym Ed. 2007;18(3):241–268. doi:10.1163/156856207779996931
24. Pi J, Yang F, Jin H, et al. Selenium nanoparticles induced membrane bio-mechanical property changes in MCF-7 cells by disturbing membrane molecules and F-actin. Bioorganic Med Chem Lett. 2013;23(23):6296–6303. doi:10.1016/j.bmcl.2013.09.078
25. Kermanizadeh A, Chauché C, Brown DM, Loft S, Møller P. The role of intracellular redox imbalance in nanomaterial induced cellular damage and genotoxicity: a review. Environ Mol Mutagen. 2015;56(2):111–124. doi:10.1002/em.21926
26. Lee KH, Jeong D. Bimodal actions of selenium essential for antioxidant and toxic pro-oxidant activities: the selenium paradox (Review). Mol Med Rep. 2012;5(2):299–304. doi:10.3892/mmr.2011.651
27. Forceville X. Seleno-enzymes and seleno-compounds: the two faces of selenium. Crit Care. 2006;10(6):180–181. doi:10.1186/cc5109
28. Brozmanová J, Mániková D, Vlčková V, Chovanec M. Selenium: a double-edged sword for defense and offence in cancer. Arch Toxicol. 2010;84(12):919–938. doi:10.1007/s00204-010-0595-8
29. Letavayová L, Vlčková V, Brozmanová J. Selenium: from cancer prevention to DNA damage. Toxicology. 2006;227(1–2):1–14. doi:10.1016/j.tox.2006.07.017
30. Chen T, Wong YS. Selenocystine induces reactive oxygen species-mediated apoptosis in human cancer cells. Biomed Pharmacother. 2009;63(2):105–113. doi:10.1016/j.biopha.2008.03.009
31. Liao S, Shi D, Clemons-Chevis CL, et al. Protective role of selenium on aflatoxin B1-induced hepatic dysfunction and apoptosis of liver in ducklings. Biol Trace Elem Res. 2014;162(1–3):296–301. doi:10.1007/s12011-014-0131-4
32. Amin KA, Hashem KS, Alshehri FS, Awad ST, Hassan MS. Antioxidant and hepatoprotective efficiency of selenium nanoparticles against acetaminophen-induced hepatic damage. Biol Trace Elem Res. 2017;175(1):136–145. doi:10.1007/s12011-016-0748-6
33. Frisk P, Yaqob A, Lindh U. Indications of selenium protection against cadmium toxicity in cultured K-562 cells. Sci Total Environ. 2002;296(1–3):189–197. doi:10.1016/S0048-9697(02)00080-3
34. Chen X, Zhu YH, Cheng XY, Zhang ZW, Xu SW. The protection of selenium against cadmium-induced cytotoxicity via the heat shock protein pathway in chicken splenic lymphocytes. Molecules. 2012;17(12):14565–14572. doi:10.3390/molecules171214565
35. Müller TE, Nunes ME, Menezes CC, et al. Sodium selenite prevents paraquat-induced neurotoxicity in zebrafish. Mol Neurobiol. 2017:1–14. doi:10.1007/s12035-017-0441-6.
36. Rzymski T, Milani M, Pike L, et al. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene. 2010;29(31):4424–4435. doi:10.1038/onc.2010.191
37. Sciarretta S, Zhai P, Shao D, et al. Activation of NADPH oxidase 4 in the endoplasmic reticulum promotes cardiomyocyte autophagy and survival during energy stress through the protein kinase RNA-activated-like endoplasmic reticulum kinase/eukaryotic initiation factor 2?/activating transcript. Circ Res. 2013;113(11):1253–1264. doi:10.1161/CIRCRESAHA.113.301787
38. Rapti K, Diokmetzidou A, Kloukina I, et al. Opposite effects of catalase and MnSOD ectopic expression on stress induced defects and mortality in the desmin deficient cardiomyopathy model. Free Radic Biol Med. 2017;110:206–218. doi:10.1016/j.freeradbiomed.2017.06.010
39. Rivera RE, Christensen VL, Edens FW, Wineland MJ. Influence of selenium on heat shock protein 70 expression in heat stressed turkey embryos (Meleagris gallopavo). Comp Biochem Physiol - a Mol Integr Physiol. 2005;142(4):427–432. doi:10.1016/j.cbpa.2005.09.006
40. Yan LJ. Pathogenesis of chronic hyperglycemia: from reductive stress to oxidative stress. J Diabetes Res. 2014;2014:1–11. doi:10.1155/2014/137919
41. Narasimhan M, Rajasekaran NS. Reductive potential – a savior turns stressor in protein aggregation cardiomyopathy. Biochim Biophys Acta - Mol Basis Dis. 2015;1852(1):53–60. doi:10.1016/j.bbadis.2014.11.010
42. Handy DE, Loscalzo J. Responses to reductive stress in the cardiovascular system. Free Radic Biol Med. 2017;109:114–124. doi:10.1016/j.freeradbiomed.2016.12.006
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.