Back to Journals » Drug Design, Development and Therapy » Volume 10
Role of cytochrome P450 2D6 genetic polymorphism in carvedilol hydroxylation in vitro
Authors Wang Z , Wang L, Xu R, Zhan Y, Huang C, Dai D, Cai J , Hu G
Received 10 February 2016
Accepted for publication 31 March 2016
Published 8 June 2016 Volume 2016:10 Pages 1909—1916
DOI https://doi.org/10.2147/DDDT.S106175
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Wei Duan
Zhe Wang,1,* Li Wang,2,3,* Ren-ai Xu,4 Yun-yun Zhan,2 Cheng-ke Huang,1 Da-peng Dai,5 Jian-ping Cai,5 Guo-xin Hu2
1Department of Pharmacy, The Second Affiliated Hospital & Yuying Children’s Hospital of Wenzhou Medical University, 2Department of Pharmacology, School of Pharmacy, Wenzhou Medical University, Wenzhou, 3Department of Pharmacy, Women’s Hospital, School of Medicine, Zhejiang University, Hangzhou, 4Department of Pharmacy, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, 5The Key Laboratory of Geriatrics, Beijing Hospital & Beijing Institute of Geriatrics, Ministry of Health, Beijing, People’s Republic of China
*These authors contributed equally to this work
Abstract: Cytochrome P450 2D6 (CYP2D6) is a highly polymorphic enzyme that catalyzes the metabolism of a great number of therapeutic drugs. Up to now, >100 allelic variants of CYP2D6 have been reported. Recently, we identified 22 novel variants in the Chinese population in these variants. The purpose of this study was to examine the enzymatic activity of the variants toward the CYP2D6 substrate carvedilol in vitro. The CYP2D6 proteins, including CYP2D6.1 (wild type), CYP2D6.2, CYP2D6.10, and 22 other novel CYP2D6 variants, were expressed from insect microsomes and incubated with carvedilol ranging from 1.0 µM to 50 µM at 37°C for 30 minutes. After termination, the carvedilol metabolites were extracted and detected using ultra-performance liquid chromatography tandem mass-spectrometry. Among the 24 CYP2D6 variants, CYP2D6.92 and CYP2D6.96 were catalytically inactive and the remaining 22 variants exhibited significantly decreased intrinsic clearance values (ranging from ~25% to 95%) compared with CYP2D6.1. The present data in vitro suggest that the newly found variants significantly reduced catalytic activities compared with CYP2D6.1. Given that CYP2D6 protein activities could affect carvedilol plasma levels, these findings are greatly relevant to personalized medicine.
Keywords: CYP2D6, carvedilol, allelic variant, catalytic activity
Introduction
Carvedilol is a β1-, β2-, and α1-adrenoreceptor blocker drug with antioxidant and antiproliferative effects. It was indicated for the treatment of hypertension, stable angina pectoris, and congestive heart failure.1,2 Cytochrome P450 2D6 (CYP2D6) is considered as a key component of carvedilol elimination through hydroxylation in the liver.3 Two hydroxy metabolites were primarily metabolized from carvedilol: O-desmethyl metabolite (CYP3A4) and hydroxyphenyl carvedilol (CYP2D6), including 4′-hydroxyphenyl carvedilol (4′-OHC) and 5′-hydroxyphenyl carvedilol (5′-OHC; Figure 1). 4′-OHC and 5′-OHC were the major metabolites, which were mainly metabolized by CYP2D6.3–6
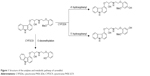  | Figure 1 Structure of the analytes and metabolic pathway of carvedilol. |
CYP2D6, a highly polymorphic enzyme, constitutes only a small percentage of the CYP450 proteins (<2%); however, it is involved in the metabolism of ~25% of clinical therapeutic drugs, including antidepressants, antitussives, β-adrenergic antagonists, antipsychotic agents, opioid drugs, and antiarrhythmic agents.7–11 CYP2D6 has extensive genetic polymorphisms with at least 105 allelic variants (http://www.cypalleles.ki.se/cyp2d6.htm). Such variability may lead to failure of treatment in carriers of alleles that results in high CYP2D6 activity. Conversely, high risk of toxicity may be associated with low-activity genotypes.12,13 Thus, the CYP2D6 polymorphism is an important determinant of carvedilol metabolism. Meanwhile, according to the activity of CYP2D6 protein in the allelic variation, it could distinguish categories of carvedilol metabolizers into four phenotypically: ultrarapid metabolizers, extensive metabolizers, intermediate metabolizers, and poor metabolizers (PMs).3,14 Poor CYP2D6 activity in PMs significantly decreases the metabolism of carvedilol, leading to higher carvedilol plasma exposure and sequentially causing high risk of toxicity.15 As mentioned in the previous report, the healthy PM subjects exerted significantly lower systolic blood pressure in the treatment of carvedilol compared with the wild type subjects.3
Previously, 22 novel nonsynonymous mutated sites among the Chinese population were found. However, the functional effects of these mutations on the metabolism of carvedilol are unclear. In the present study, we determined the two metabolites of carvedilol generated by CYP2D6 enzyme among CYP2D6.1, CYP2D6.2, CYP2D6.10, and 22 novel variants in the microsomes expressed in the insect cell and analyzed the catalytic activities of the allelic variants. And we hope this result can provide valuable information relevant to CYP2D6 genetic polymorphisms in carvedilol metabolism and the personalized oral dosing for further studies in clinic.
Materials and methods
Materials
Baculosomes coexpressing human CYP2D6 and NADPH-cytochrome P450 oxidoreductase (OR) or cytochrome b5 and OR were purchased from BD Gentest (Woburn, MA, USA). Carvedilol, 4′-OHC, 5′-OHC, metoprolol, and NADPH were obtained from Sigma-Aldrich Co. (St Louis, MO, USA). High-pressure liquid chromatography grade was obtained from Thermo Fisher Scientific (Waltham, MA, USA). Other reagents and solvents used were of analytical grade or the highest commercially available grade.
Methods
Expression of CYP2D6 variants in insect cell microsomes
Baculosomes coexpressing human CYP2D6 enzyme and NADPH-CYP450 OR were prepared and determined using methods previously described.16 Briefly, dual expression vector pFastBac-OR-CYP2D6 was packaged into baculovirus according to the manufacturer’s procedure. Then, the virus was used for the infection of Sf21 insect cells to coexpress the CYP2D6 variants and OR protein simultaneously. Three days after infection, the cells were sonicated and centrifuged to get the microsomes containing CYP2D6 variants. To quantify and clarify the correct expression of CYP2D6 variants, the recombinant CYP2D6 baculosome reagent purchased from BD Gentest was used as the microsomal protein standard in the immunoblotting analysis.16
Incubation procedure and analysis of CYP2D6 enzyme activity
Incubation mixtures were prepared in a total volume of 200 μL as follows: 5 pmol CYP2D6.1 or 10 pmol other variants, 5 pmol purified cytochrome b5, 10 mM phosphate-buffered saline (pH 7.4), 1 mM NADPH, and carvedilol final concentration ranging from 1 μM to 50 μM. The total concentration of methanol was <0.4%. In all, 20 μL of 10 mM NADPH was added to start the reaction at 37°C in a final volume of 200 μL. There was a 5-minute preincubation period at 37°C before the 30-minute reaction was initiated by adding the NADPH. Incubations were terminated by the addition of 400 μL cold acetonitrile and 40 μL metoprolol (2 μg/mL in methanol solution) as an internal standard. After being vortexed for 2 minutes, the samples were centrifuged at 10,000× g for 10 minutes. Then 2 μL of the supernatants was subjected to ultra-performance liquid chromatography tandem mass-spectrometry (UPLC-MS/MS) assay. The incubations were completed in triplicate, and the data are expressed as the mean ± SD of three experiments.
Chromatographic and mass detection conditions
Quantitative analysis was conducted with a Waters Acquity UPLC System (Waters Corp., Milford, MA, USA) equipped with Xevo TQD triple quadrupole mass spectrometer (Waters Corp.) and electrospray ionization in the positive ionization mode. The separation of analytes was performed on an UPLC BEH C18 column (100×2.1 mm, 1.7 μm; Waters Corp.).
The mobile phase consisted of 0.1% formic acid in water (solvent A) and acetonitrile (solvent B) at an isocratic flow rate of 0.4 mL/min with the following gradient: 25% B (0–0.3 minutes), 25%–36% B (0.3–3 minutes), 36%–95% B (3–3.5 minutes), 95% B (3.5–4.5 minutes), and 95%–25% B (4.5–4.6 minutes). Before the next injection, re-equilibration was performed for a subsequent time of 1.0 minutes. The injection volume was 2 μL. Under these conditions, the retention times of metoprolol, 4′-OHC, 5′-OHC, and carvedilol were 1.28 minutes, 2.24 minutes, 2.39 minutes, and 3.73 minutes, respectively.
The precursors to product ion transitions were monitored at m/z 407.1→99.2, 423.03→99.73, 423.1→99.73, and 268.09→115.92 for carvedilol, 4′-OHC, 5′-OHC, and metoprolol, respectively. The optimal mass parameters for the analytes and internal standards were as follows: capillary voltage, 2.5 kV; source temperature, 150°C; and desolvation temperature, 350°C. The pressure of argon used as collision activation dissociation gas was 0.137 Pa. The optimum values for compound-dependent parameters such as cone voltage and collision energy were set at 40 V and 16 eV for carvedilol, 50 V and 30 eV for 4′-OHC, 50 V and 30 eV for 5′-OHC, and 40 V and 16 eV for metoprolol, respectively. Quadrupoles 1 and 3 were maintained at unit mass resolution, and the dwell time was set at 100 ms. MassLynx Software Version 4.1 (Waters Corp., Milford, MA, USA) was used to control all parameters of UPLC-MS/MS.
Statistical analysis
Michaelis–Menten analysis was estimated by nonlinear regression curve fitting utilizing GraphPad Prism Version 5 (GraphPad Software, Inc., La Jolla, CA, USA). All kinetic data were expressed as the mean ± SD of three microsomal preparations. Statistical analyses were analyzed with IBM SPSS Statistics (Version 19.0; IBM Corporation, Armonk, NY, USA). The statistical analyses were evaluated by one-way analysis of variance for intergroup comparison. Dunnet’s multiple-range test was used for the catalytic activity data between CYP2D6.1 and other variants. P-value <0.05 was considered to be statistically significant.
Results
Michaelis–Menten plots of 4′-hydroxylation and 5′-hydroxylation for each of the CYP2D6 variants are shown in Figures 2 and 3, respectively, and the corresponding kinetic parameters of 4′-hydroxylation and 5′-hydroxylation for carvedilol are summarized in Tables 1 and 2, respectively. The values of intrinsic clearance (Vmax/Km) for carvedilol 4′-hydroxylation and 5′-hydroxylation were significantly decreased in all the tested allelic variants.
CYP2D6.92 and CYP2D6.96 did not show any enzyme activity toward carvedilol 4′-hydroxylation. CYP2D6.89, E215K, and R440C exhibited the Vmax values, which were 2.24-, 1.68-, and 1.35-fold higher than CYP2D6.1, respectively; however, Km values of the three variants increased more apparently than Vmax values (3.01-, 7.42-, and 2.94-fold higher than CYP2D6.1, respectively). Therefore, CYP2D6.89, E215K, and R440C showed intrinsic clearance 1.34-, 4.41-, and 2.17-fold lower compared with that of CYP2D6.1, respectively.
Nine of the 24 defective alleles (CYP2D6.87, CYP2D6.91, CYP2D6.95, CYP2D6.97, R25Q, E215K, V327M, V342M, and R497C) exerted similar values of intrinsic clearance to the typical defective allele CYP2D6.2 (Table 1). CYP2D6.93 showed comparable values to another typical defective allele CYP2D6.10 that presented 22.94-fold lower intrinsic clearance value than that of CYP2D6.1. The remaining ten variants (CYP2D6.88, CYP2D6.89, CYP2D6.90, CYP2D6.94, CYP2D6.98, F164L, F219S, D336N, R344Q, and R440C) also exhibited significantly reduced intrinsic clearance values compared with CYP2D6.1.
The values of intrinsic clearance (Vmax/Km) for carvedilol 4′-hydroxylation were significantly decreased in all the allelic variants in the present research. Carvedilol 5′-hydroxylation exhibited similar results to that of carvedilol 4′-hydroxylation. Michaelis–Menten curves for each of the CYP2D6 variants are shown in Figure 3, and the kinetic parameters of each CYP2D6 enzymatic activity are summarized in Table 2.
Discussion
CYP2D6, an important enzyme of CYP450 subfamily in clinic, metabolized a certain number of medicines. CYP2D6 was primarily responsible for carvedilol metabolism, and the polymorphisms play an important role in the wide interindividual variation in the plasma levels.17 In this study, we observed the functional alterations in CYP2D6 allelic variants toward carvedilol hydroxylation. To identify the effects of 24 CYP2D6 variants on carvedilol hydroxylation, the catalytic activities of CYP2D6.1 and 24 other CYP2D6 variant proteins expressed in insect cells were analyzed. All the 24 allelic variants exhibited significantly reduced intrinsic clearance values compared with the CYP2D6.1.
The CYP2D6.2 and CYP2D6.10, two typical defective genotypes, undergo the most research concerning their frequencies and catalytic activities. In this study, CYP2D6.2 exhibited lower catalytic activity than CYP2D6.1 in carvedilol 4′-hydroxylation (69.59% of CYP2D6.1) and 5′-hydroxylation (73.20% of CYP2D6.1), respectively. CYP2D6.10, occurring in ~50% of individuals in oriental populations, was one of the most widely studied CYPs in relation to its genetic polymorphisms.18,19 This variant contains C/T 188 and G/C 4268 mutations and results in the crucial amino acid substitutions of Pro34Ser and Ser486Thr.11,20 A previous report indicated that the intrinsic clearance value of CYP2D6.10 in clinical medicine was significantly decreased, including bufuralol, dextromethorphan, debrisoquine, atomoxetine, tramadol, and codeine.3 In the present research, CYP2D6.10 was associated with smaller values of Vmax in carvedilol 4′-hydroxylation (4.36% of CYP2D6.1) and 5′-hydroxylation (3.56% of CYP2D6.1), and thus caused lower clearance value compared with CYP2D6.1. This observation indicates that the metabolism in CYP2D6.10 allele carriers may be saturated even at low dose compared with the noncarriers. Similar to CYP2D6.10, CYP2D6.93 exhibited low clearance value in carvedilol 4′-hydroxylation (7.19% of CYP2D6.1) and 5′-hydroxylation (5.48% of CYP2D6.1) with significant alteration in Vmax.
CYP2D6.92 (218Frameshift) and CYP2D6.96 (Gln424STOP) did not show any catalytic activities even at the substrate concentration of 50 μM. Therefore, the two inactive variants can be classified as poor metabolism phenotypes. Similarly, CYP2D6.20 (211Frameshift) and CYP2D6.8 (Gly169STOP) also did not exhibit any catalytic activity. It is perhaps reasonable to infer that enzymatic function abolished may be associated with the frameshift or the premature termination in the protein translation process.15
The Glu216 residue was located in the F helix, which was essential for the active site cavity.21,22 Its replacement with a neutral or basic residue could significantly decrease the binding of amine substrates and sequentially caused reduction or abolishment in enzymatic activity.23 The present result further showed that E215K and F219S displayed low activity similar to that of CYP2D6.2. Furthermore, the results reinforce previous observations showing that mutations 1984G>A (p.E215K) and 1997T>C (p.F219S) located in the F helix would affect the activity of CYP2D6.22
Meanwhile, the new allelic isoform R25Q also decreased significantly in catalytic activity toward carvedilol. A previous study showed that CYP2D6.47, which contains R25W, P34S, and S486T substitutions, reveals poor enzymatic activities similar to that of CYP2D6.10 (P34S and S486T).10 Thus, it could be inferred that Arg25 plays role in halting the transfer signal and involves direct decrease of the enzyme activity.10
In the previous functional prediction of the 22 novel variants, CYP2D6 protein activity might be reduced in eight variants (L142S, C161S, F219S, T249P, 424STOP, R440C, H463D, and R497C).22 In the present study, T249P (CYP2D6.93) showed decreased catalytic activity that was similar to CYP2D6.10. Meanwhile, C161S (CYP2D6.91) and R497C exerted decreased catalytic activity that was similar to CYP2D6.2. 424STOP (CYP2D6.96) exhibited a deficiency of metabolic activity. The other variants (L142S (CYP2D6.89), F219S, R440C, and H463D (CYP2D6.98)) exhibited lower catalytic activity compared with CYP2D6.1, but higher to CYP2D6.2. It provided the evidence that an amino acid substitution will impact on the structure and function and subsequently affect the activity of CYP2D6 enzyme. Meanwhile, single nucleotide polymorphisms could be primarily responsible for the function of CYP2D6 enzyme activity. For example, 100C>T and 1039C>T (CYP2D6.94) and 2850C>A (CYP2D6.91) existed in the 5′ and 3′ ends of the CYP2D6 gene, respectively.22
In the present study, CYP2D6.1 and 24 other CYP2D6 alleles were functionally assessed, including 22 recently detected nonsynonymous mutations found in Chinese Han populations. Importantly, the results in this study demonstrated that all the variants tested significantly reduce the metabolic activities toward the substrate carvedilol. Further in vivo research should confirm whether defective allele carriers require low oral doses of carvedilol or other drugs metabolized on CYP2D6.
Acknowledgment
The work was supported by a grant from the National Health and Family Planning Commission of the People’s Republic of China (201302008).
Author contributions
ZW carried out the in vitro studies and drafted the manuscript. LW performed in vitro studies. YYZ participated in the in vitro studies. DPD participated in the in vitro studies. JPC conceived the study, participated in the design of the study, and participated in the statistical analysis. GXH conceived the study and performed the design coordination. All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work. All authors read and approved the final manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Beattie K, Phadke G, Novakovic J. Chapter four – carvedilol. In: Harry GB, editor. Profiles of Drug Substances, Excipients and Related Methodology. Cambridge: Academic Press; 2013:113–157. | ||
Stanley AJ, Dickson S, Hayes PC, et al. Multicentre randomised controlled study comparing carvedilol with variceal band ligation in the prevention of variceal rebleeding. J Hepatol. 2014;61(5):1014–1019. | ||
Zhou S-F. Polymorphism of human cytochrome P450 2D6 and its clinical significance. Clin Pharmacokinet. 2009;48(12):761–804. | ||
Giessmann T, Modess C, Hecker U, et al. CYP2D6 genotype and induction of intestinal drug transporters by rifampin predict presystemic clearance of carvedilol in healthy subjects. Clin Pharmacol Ther. 2004;75(3):213–222. | ||
Pan P-P, Weng Q-H, Zhou C-J, et al. The role of CYP2C9 genetic polymorphism in carvedilol O-desmethylation in vitro. Eur J Drug Metab Pharmacokinet. 2014;41(1):1–8. | ||
Shihmanter R, Nulman I, Goland S, et al. Variation in the CYP2D6 genotype is not associated with carvedilol dose changes in patients with heart failure. J Clin Pharm Ther. 2014;39(4):432–438. | ||
Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther. 2007;116(3):496–526. | ||
Ingelman-Sundberg M. Pharmacogenetics of cytochrome P450 and its applications in drug therapy: the past, present and future. Trends Pharmacol Sci. 2004;25(4):193–200. | ||
Zanger UM, Raimundo S, Eichelbaum M. Cytochrome P450 2D6: overview and update on pharmacology, genetics, biochemistry. Naunyn Schmiedebergs Arch Pharmacol. 2004;369(1):23–37. | ||
Wang B, Yang LP, Zhang XZ, Huang SQ, Bartlam M, Zhou SF. New insights into the structural characteristics and functional relevance of the human cytochrome P450 2D6 enzyme. Drug Metab Rev. 2009;41(4):573–643. | ||
Li Q, Wang R, Guo Y, Wen S, Xu L, Wang S. Relationship of CYP2D6 genetic polymorphisms and the pharmacokinetics of tramadol in Chinese volunteers. J Clin Pharm Ther. 2010;35(2):239–247. | ||
Dorado P, Heras N, Machín E, Hernández F, Teran E, Llerena A. CYP2D6 genotype and dextromethorphan hydroxylation phenotype in an Ecuadorian population. Eur J Clin Pharmacol. 2012;68(5):637–644. | ||
Llerena A, Dorado P, Peñas-Lledó EM. Pharmacogenetics of debrisoquine and its use as a marker for CYP2D6 hydroxylation capacity. Pharmacogenomics. 2009;10(1):17–28. | ||
Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 2005;5(1):6–13. | ||
Zhou S-F, Liu J-P, Chowbay B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab Rev. 2009;41(2):89–295. | ||
Cai J, Dai DP, Geng PW, et al. Effects of 22 novel CYP2D6 variants found in the Chinese population on the bufuralol and dextromethorphan metabolisms in vitro. Basic Clin Pharmacol Toxicol. 2016;118(3):190–199. | ||
Oldham HG, Clarke SE. In vitro identification of the human cytochrome P450 enzymes involved in the metabolism of R(+)- and S(−)-carvedilol. Drug Metab Dispos. 1997;25(8):970–977. | ||
Saruwatari J, Nakashima H, Tsuchimine S, Nishimura M, Ogusu N, Yasui-Furukori N. Possible impact of the CYP2D6*10 polymorphism on the nonlinear pharmacokinetic parameter estimates of paroxetine in Japanese patients with major depressive disorders. Pharmgenomics Pers Med. 2014;7:121–127. | ||
Teh LK, Bertilsson L. Pharmacogenomics of CYP2D6: molecular genetics, interethnic differences and clinical importance. Drug Metab Pharmacokinet. 2012;27(1):55–67. | ||
Johansson I, Oscarson M, Yue QY, Bertilsson L, Sjöqvist F, Ingelman-Sundberg M. Genetic analysis of the Chinese cytochrome P4502D locus: characterization of variant CYP2D6 genes present in subjects with diminished capacity for debrisoquine hydroxylation. Mol Pharmacol. 1994;46(3):452–459. | ||
Rowland P, Blaney FE, Smyth MG, et al. Crystal structure of human cytochrome P450 2D6. J Biol Chem. 2006;281(11):7614–7622. | ||
Qian JC, Xu XM, Hu GX, et al. Genetic variations of human CYP2D6 in the Chinese Han population. Pharmacogenomics. 2013;14(14):1731–1743. | ||
Guengerich FP, Hanna IH, Martin MV, Gillam EM. Role of glutamic acid 216 in cytochrome P450 2D6 substrate binding and catalysis. Biochemistry. 2003;42(5):1245–1253. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




