Back to Journals » Clinical Interventions in Aging » Volume 14
Risk Factors And Epigenetic Markers Of Left Ventricular Diastolic Dysfunction With Preserved Ejection Fraction In A Community-Based Elderly Chinese Population
Authors Wang W, Zhang Y , Wang R , Shrestha Y, Xu Y, Peng L, Zhang J, Li J, Zhang L
Received 17 June 2019
Accepted for publication 19 September 2019
Published 4 October 2019 Volume 2019:14 Pages 1719—1728
DOI https://doi.org/10.2147/CIA.S219748
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Zhi-Ying Wu
Wei Wang,1,2 Yi Zhang,3 Runzi Wang,1,2 Yeshaswi Shrestha,1,2 Yawei Xu,3 Luying Peng,1,4,5 Jie Zhang,1,2 Jue Li,1,2 Lijuan Zhang1,2
1Key Laboratory of Arrhythmias of the Ministry of Education, Tongji University School of Medicine, Shanghai 200092, People’s Republic of China; 2Institute of Clinical Epidemiology and Evidence-Based Medicine, Tongji University School of Medicine, Shanghai 200092, People’s Republic of China; 3Department of Cardiology, Shanghai Tenth People’s Hospital, Tongji University School of Medicine, Shanghai 200072, People’s Republic of China; 4Department of Pathology and Pathophysiology, Tongji University School of Medicine, Shanghai 200092, People’s Republic of China; 5Research Center for Translational Medicine, Shanghai East Hospital, Tongji University School of Medicine, Shanghai 200120, People’s Republic of China
Correspondence: Jue Li; Lijuan Zhang
Key Laboratory of Arrhythmias of the Ministry of Education, Tongji University School of Medicine, Shanghai 200092, People’s Republic of China
Tel/fax +86 21 65985195
Email [email protected]; [email protected]
Purpose: Left ventricular diastolic dysfunction with preserved ejection fraction (LVDD-PEF) is an early-stage manifestation but poorly understood in the process of heart failure. This study was designed to investigate risk factors and epigenetic markers for predicting LVDD-PEF.
Patients and methods: A community-based study in 1568 residents over 65 years was conducted in Shanghai, People’s Republic of China, from June 2014 to August 2015. Echocardiography was performed to diagnose LVDD-PEF. DNA methylation by whole-genome bisulfite sequencing was used to determine those potential epigenetic markers contributing to LVDD-PEF.
Results: A total of 177 participants (11.3%) were diagnosed with LVDD-PEF, and higher prevalence in females than in males (15.0% vs 6.5%, P<0.001). Multivariate logistic regression analysis indicated that female sex (OR 2.46, 95% CI 1.47–4.13), body mass index (BMI) (OR 1.09, 95% CI 1.04–1.14), pulse pressure (PP) (OR 1.03, 95% CI 1.01–1.05) and carotid intima-media thickness (CIMT) (OR 4.20, 95% CI 1.40–12.55) showed a significant association with LVDD-PEF. Overall, 638 CpG sites were differentially methylated in LVDD-PEF group compared to non-LVDD-PEF group (P<0.001); 242 sites were significantly hypermethylated (covering 238 genes) and 396 sites were significantly hypomethylated (covering 265 genes).
Conclusion: Our findings found female, BMI, PP, and CIMT were independent predictors for LVDD-PEF in the community-dwelling elderly population. Regulation of DNA methylation might play a crucial role for LVDD-PEF.
Keywords: left ventricular diastolic dysfunction, DNA methylation, risk factor
Introduction
Heart failure (HF) is a complex pathophysiological syndrome with severe morbidity and mortality. Its prevalence varies from 6% to 10% in those aged over 65 years and increases substantially with aging. Heart failure leads to a lethal condition, with an annual mortality of about 17% in women and 21% in men.1 Up to 50% of patients diagnosed with end-stage HF will die within 1 year. Evidence indicates that early diagnosis and effective intervention might be effective measures to slow the progression of HF and reduce those adverse cardiovascular (CV) events.2 European Society of Cardiology guidelines (2016) on HF indicate that among people over 65 years presenting to primary care with breathlessness on exertion, one in six will have unrecognized HF with preserved ejection fraction (HFpEF).3 Left ventricular diastolic dysfunction with preserved ejection fraction (LVDD-PEF) is recognized frequently as the earlier alteration of HF, classified in those asymptomatic individuals with a left ventricular ejection fraction (LVEF) over 50%. This syndrome regularly results in impaired filling function with or without reduced restoring forces, which increase cardiac filling pressures.4 LVDD-PEF was very common in the community-based elderly population, with a median prevalence of 36.0% (15.8–52.8%), and it could rise steeply with age.5 Recent studies demonstrated that patients with worsened diastolic dysfunction had substantial risk for HF, even risk of early death.6,7 Defined by the American College of Cardiology/American Heart Association, LVDD-PEF almost stays in stage B of HF.8 Currently, LVDD-PEF patients have less predictive biomarkers to better stratify individuals at risk.9 Clinical HF guidelines emphasis the importance of early identification for the cardiac dysfunction to prevent the cardiac pathological conditions.8 The last decades have witnessed the huge amount of studies linking cardiac disorders to epigenetic regulation that governing potential pathogenesis. Application of human genetic technologies has accelerated the advancement in our comprehension of the role of epigenetics and risk factors involved in the pathogenesis of cardiovascular disease (CVD). Epigenetic modifications can lead to exacerbate the stage of HF with consequences on cardiac remodeling and preceding cardiac dysfunction.10 Previous studies suggested that DNA methylation was associated with hypertrophy and can reduce cardiac contractility.9,11 Therefore, current evidence supports the importance of DNA methylation alteration in the process of HF. In the present community-based cross-sectional study, we aimed to detect potential risk factors and epigenetic markers for LVDD-PEF in order to explore the independent predictive factors for this early-stage of HF which could act as a target for future therapeutic strategies.
Materials And Methods
An ongoing community-based investigation named the Northern Shanghai Study was performed from June 2014 to August 2015 in the northern urban area of Shanghai, People’s Republic of China (Clinical trial registration: NCT02368938). Details of the “Northern Shanghai Study” methodology have been described elsewhere.12 Briefly, elderly participants were recruited to build a CV risk score. Eligibility criteria included in the following: 1) the individual should be a long-term resident in northern Shanghai; 2) age ≥65 years old; 3) telephone in the home or easy access to one; 4) signed the informed consent voluntarily and can be followed up for a long period.
Exclusion criteria included: 1) had severe CVD (achieve IV grade of New York Heart Association); 2) previously diagnosed with end-stage of renal disease (chronic kidney disease ≥4); 3) suffered from cancer or less than 5 years of life expectancy; 4) experienced stroke within 3 months; 5) could not return for the follow-up visits and declined to participate in the study. Finally, a total of 1599 participants were enrolled in this investigation and 1568 subjects who completed both echocardiography and carotid ultrasonography detection were included in the final analysis. The study was approved by the medical ethical review committee of Tongji University and informed consent was obtained for all participants. This study was conducted in accordance with the Declaration of Helsinki.
A structured questionnaire was used to obtain basic information and medical data, including age, gender, height, weight, drinking and smoking habits, and history of hypertension, diabetes mellitus, coronary heart disease, and stroke. Venous blood samples and urine samples were obtained from fasting individuals in the morning. Biological markers, including plasma low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, plasma creatinine (PCr), urinary microalbumin and creatinine, were measured by uniformed methods in the Department of Laboratory Medicine of Shanghai Tenth People’s Hospital. Fasting plasma glucose was measured by the glucose oxidase method. Body weight (in kilograms) and body height (in meters) were measured by a well-trained clinician. Participants wore light clothes and removed shoes for weight measurements. Body mass index (BMI) was calculated as weight (kg) divided by height squared (m2). Blood pressure (BP) was measured three times every 5 mins in the morning at the room temperature in the sitting position, after resting for 10 mins using a mercury sphygmomanometer. Hypertension was defined as brachial SBP ≥140mmHg or DBP ≥90 mmHg or the use of antihypertensive agents or a previous history of hypertension.13 The average of the three readings was regarded as participants’ BP and was used for further analysis. Pulse pressure (PP) was calculated as the difference between SBP and DBP. Echocardiography and carotid ultrasonography were measured by a single skillful, experienced and well-trained cardiologist in the Department of Cardiology, Shanghai Tenth People’s Hospital who was blinded to participants’ clinical data using the MyLab30 Gold CV system (Esaote SpA, Genoa, Italy) followed by the American Society of Echocardiography recommendations.14 Carotid intima-media thickness (CIMT) was measured in the left common carotid artery by using carotid ultrasonography, which is on plaque-free segments – nearly 2 cm from the bifurcation. The measurement of CIMT was taken three times, and the average result was used for analysis. Left ventricular dimensions and mass were measured through transthoracic echocardiography. Left ventricular mass (LVM) was calculated from two-dimensionally guided M-mode echocardiograms. The LVEF was obtained according to Teichholz’s formula.15 Left atrium (LA) size was measured in the parasternal long-axis and apical four chamber views. Left ventricular end-diastolic diameter (LVEDd), interventricular septal (IVSd), and posterior wall thickness at end-diastole (PWTd) were measured directly which were used to calculate LVM.14 The formula is:
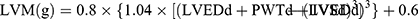
Left ventricular mass index (LVMI) was obtained by dividing the raw LVM by body surface area (BSA). In addition, left atrial volume was calculated using the ellipse model formula:
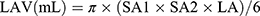
In this formula, left atrial parameters were measured including SA1, SA2 and LA. SA1 is the M-mode left atrial dimension in the parasternal short-axis view and SA2 and LA are measurements of short and long axes in the apical four chamber view at ventricular end-systole. Left atrial volume index (LAVI) was also standardized by BSA.16 Peak early diastolic transmitral flow velocity (E) and early diastolic lateral mitral annular velocity (E’) were performed by pulse-wave Doppler imaging and tissue Doppler signals in lateral septum, respectively. The ratio of E/E’ was present to evaluate LV diastolic function. Left ventricular diastolic dysfunction (LVDD) was assessed by E/E’ according to following criteria: E/E’ ≥15; or 8 < E/E’<15 with any of the conditions 1) LAVI >40 mL/m2; 2) LVMI >149 g/m2 (male); 3) LVMI >122 g/m2 (female).17,18
Confirmed LVDD-PEF patients and healthy controls were recruited for the detection of DNA methylation changes, as a part of the Northern Shanghai Study. Whole-genome bisulfite sequencing (WGBS) was performed in 15 randomized subjects (age 76.76 7.00 years, mean
7.00 years, mean SE) for the present study. After screening 177 patients with LVDD-PEF, 10 LVDD-PEF subjects were a random sample of those who were in the absence of relevant heart diseases. A total of 5 controls were randomly selected from the non-LVDD-PEF group with normal diastolic function and no evidence for other CV disorders as determined by echocardiography, matched by age and gender. No significant difference was observed for baseline characteristics between two groups using a Wilcoxon rank test.
SE) for the present study. After screening 177 patients with LVDD-PEF, 10 LVDD-PEF subjects were a random sample of those who were in the absence of relevant heart diseases. A total of 5 controls were randomly selected from the non-LVDD-PEF group with normal diastolic function and no evidence for other CV disorders as determined by echocardiography, matched by age and gender. No significant difference was observed for baseline characteristics between two groups using a Wilcoxon rank test.
For each participant enrolled in the DNA methylation exploration, whole blood was obtained after an overnight fast and immediately frozen and stored at −80°C. WGBS for DNA methylation analysis was extracted from previously frozen peripheral blood using phenol-chloroform protocol. The DNA was fragmented by sonication (Covaris) using a Bioruptor (Diagenode, Belgium) to a mean size of 250 bp, followed by blunt-ending, dA addition to 3ʹ-end and, finally, adaptor ligation (in this case of methylated adaptors to protect from bisulfite conversion), essentially according to the manufacturer’s instructions. Bisulfite conversion of genomic DNA was carried out with EZ DNA Methylation-Gold Kit (ZYMO) following the manufacturer’s protocol. Different insert size fragments were excised from the same lane of a 2% TAE agarose gel. At last, sequencing was performed using HighSeq4000 or other Illumina platforms (pipeline). The reads generated by Illumina sequencing were aligned to the reference genome using SOAP aligner. The alignment and methylation estimation was performed as described previously. Averagely 0.8 GB clean reads of each sample were generated after filtering low quality reads, N reads and adaptor sequences. After filtering, clean reads were aligned to the human reference genome (hg19) using BSMAP.19 Methylation level was determined by dividing the number of reads covering each mC by the total reads covering that cytosine. In general, CG methylation is found in both genes and repeats, and is involved in gene expression regulation.
Differentially methylated regions (DMRs) are stretches of DNA in a sample’s genome that have different patterns compared with other samples. Putative DMRs were identified by comparison of the sample1 and sample2 methylomes using windows that contained at least 5 CpG(CHG or CHH) sites with a twofold change in methylation level and Fisher test P-value ≤0.05. Two nearby DMRs would be considered interdependent and joined into one continuous DMR if the genomic region from the start of an upstream DMR to the end of a downstream DMR also had twofold methylation level differences between sample1 and sample2 with a P-value ≤0.05. Otherwise, the two DMRs were viewed as independent. After iteratively merging interdependent DMRs, the final dataset of DMRs was made up of those that were independent of each other.
Categorical and continuous variables were expressed as numbers with percentage or means±SD. Differences between LVDD-PEF and non-LVDD-PEF were analyzed by independent t-test for continuous variables or by χ2 test for categorical variables. Multiple stepwise linear regression was applied for the association of diastolic function (E/E’). Multiple stepwise logistic regression analyses were performed to investigate the associations between independent variables and LVDD-PEF both under a full model and an adjusted model with age and gender were adjusted. Statistical analysis was performed using SPSS software, version 20.0 (SPSS Inc., Chicago, IL). P-value<0.05 was considered to be statistically significant.
Gene ontology (GO), which is a major bioinformatics initiative to unify the representation of gene and gene product attributes in any organism, provides an ontology of defined terms representing gene product properties. To investigate pathways and processes that may be subject to epigenetic variation in association with DMRs, DMRs-related genes both obtained from all samples were analyzed using GO and pathway analysis. All DMR-related genes were mapped to GO terms in the database (http://www.geneontology.org/), calculating gene numbers for every term and using the hypergeometric test to find significantly enriched GO terms in the input list of DMR-related genes, based on “GO: TermFinder” (http://www.yeastgenome.org/help/analyze/go-term-finder). The calculated P-value goes through bonferroni correction,20 taking corrected P-value ≤0.05 as a threshold. GO terms fulfilling this condition are defined as significantly enriched GO terms in DMR-related genes.
Pathway-based analysis helps to further understand genes biological functions. KEGG21(Kyoto Encyclopedia of Genes and Genomes) is used to better perform pathway enrichment analysis of DMR-related genes which can identify significantly enriched metabolic pathways or signal transduction pathways in DMR-related genes comparing the whole-genome background. The formula used for the calculation is the same as that in GO analysis. Significantly higher DNA methylation levels between the cases and controls were identified using a Wilcoxon rank test. P-values were corrected for multiple testing according to the Bonferroni method. P≤0.001 was considered significant.
Results
Baseline characteristics of subjects are presented in Table 1. Of the whole study population, the average age was 71.9 6.1 years; 878 (56.0%) participants were women; average BMI was 24.23
6.1 years; 878 (56.0%) participants were women; average BMI was 24.23 3.46 kg/m2. Of 1568 participants, 177 (11.3%) subjects were diagnosed with LVDD-PEF. About half of the population (819, 52.2%) had hypertension; 33.9% of participants (531) had CVD history; 19.5% of participants (306) had diabetes mellitus. The comparison between LVDD-PEF and non-LVDD-PEF indicated that female sex, BMI, SBP, PP, history of hypertension and CVD, and echocardiographic parameters in participants with LVDD-PEF were significantly higher than those without LVDD-PEF (all P<0.05).
3.46 kg/m2. Of 1568 participants, 177 (11.3%) subjects were diagnosed with LVDD-PEF. About half of the population (819, 52.2%) had hypertension; 33.9% of participants (531) had CVD history; 19.5% of participants (306) had diabetes mellitus. The comparison between LVDD-PEF and non-LVDD-PEF indicated that female sex, BMI, SBP, PP, history of hypertension and CVD, and echocardiographic parameters in participants with LVDD-PEF were significantly higher than those without LVDD-PEF (all P<0.05).
 |
Table 1 Baseline Characteristics Of Participants |
Considering the risk factors related to LVDD, stepwise multiple linear regression was performed to analyze the independent issues of E/E’ and the results indicated that E/E’ was significantly associated with BMI (0.067±0.028, P=0.016), PP (0.053±0.012, P<0.001) and CIMT (1.385±0.615, P=0.027) (Table 2).
 |
Table 2 Determinants Of E/E’ By Multiple Linear Regression Analysis |
Age and gender were highly predictive of LVDD-PEF. Significant association with LVDD-PEF included BMI (OR 1.10; 95% CI 1.05–1.15), SBP (OR 1.02; 95% CI 1.01–1.03), PP (OR 1.03; 95% CI 1.02–1.04), CIMT (OR 4.64; 95% CI 1.67–12.93), plaque in right carotid artery (OR 0.71; 95% CI 0.52–0.99) and history of hypertension (OR 1.98; 95% CI 1.42–2.76) after adjustment by age and gender (Table 3). In a multivariate logistic analysis model (Table 4), BMI (OR 1.09; 95% CI 1.04–1.14), PP (OR 1.03; 95% CI 1.01–1.05) and CIMT (OR 4.20; 95% CI 1.40–12.55) still the independent predictors of LVDD-PEF, and effects of SBP, plaque in right carotid artery and history of hypertension were attenuated (all P>0.05).
 |
Table 3 Age- And Gender-Adjusted Multivariate Logistic Analyses |
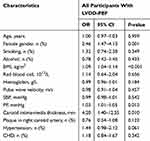 |
Table 4 Independent Risk Factors Of LVDD-PEF According To Multivariate Logistic Analysis |
Ten participants with LVDD-PEF and 5 participants without LVDD-PEF were further included in the DNA methylation analysis. A total of 639 sites (covering 580 genes) were significantly differentially methylated between two groups (Wilcoxon rank test, P<0.001). The extent of DNA methylation was significantly increased at 242 sites in 238 genes, while 396 sites were significantly hypo-methylated in 265 genes in LVDD-PEF group compared with controls. We focused on the differentially methylated genes in the upstream region since hyper-methylation in promoters might cause alteration of gene expression. The heatmap showed the graphical representation of differentially hyper and hypomethylated genes in LVDD-PEF subjects and controls (Figure 1). By performing heatmap cluster analysis, 8 hypermethylated (HIST1H2BK, HAUS8, RPUSD1, C3orf14, ALDH4A1, CDH1, CCN and RGS10) and 17 hypomethylated (SQLE, SERPINA1, H2AFJ, GPRC5C, SLC18A2, CRACR2A, RPP38, TMEM5, SNRPE, SLC4A3, DLX6, ACTG2, NXN, RAPGEF5, IFI6, SCD5 and PPP2R5D) genes were identified as significantly differentially methylated genes in the group of LVDD-PEF (Table 5). Three of these candidate genes, RGS10, CRACR2A and ACTG2 were further identified by searching the PubMed (NCBI) and GENECARDS database comprehensively (Table 6) which might be involved in the cardiac physiological process.
 |
Table 5 Details Of Differentially Methylated Regions Between Two Groups |
 |
Table 6 Candidate Genes Selected For Validation |
Discussion
It is critical to investigate the clinical variables and mechanisms of LVDD-PEF since the outcome and prognosis of HFpEF have been poorly understood.22,23 Both the modifiable and nonmodifiable elements contributed to the progression of LVDD-PEF. This community-based study was designed to clarify risk factors and potential epigenetic markers for predicting LVDD-PEF in Chinese elderly population. We identified 11.3% of the participants were displayed LVDD-PEF, which was less than 15.8–52.8% reported by previous studies.24–27 We also confirmed that female gender, BMI, PP and CIMT were closely correlated with the risk of LVDD-PEF. To be precise, increase in BMI, PP and CIMT would be predictive markers for the value of E/Eˊ, and they then predicted LVDD-PEF even after adjustment of those potential confounders. Similarly, Perkiömäki et al28 discovered that female gender was associated with diastolic dysfunction in the middle-aged population. BMI was also reported to be an independent predictor of LVDD (OR 1.06, 95% CI 1.04–1.08).29
As one of the early stages of HF progression, LVDD-PEF was responsible for clinical and symptomatic deterioration. Diastolic dysfunction can be induced through increased LV stiffness caused by hypertrophy and interstitial fibrosis, as well as from abnormal LV relaxation due to abnormal calcium cycling.30 In our present analysis, PP was considerably associated with LVDD-PEF. Elevated PP in elderly population could lead to increase the arterial stiffness and thicken the left ventricular walls, as the result of an increase in LVM.31 CIMT, also regarded as an early biomarker for the development of atherosclerosis, even for vascular remodelling and ageing. Arterial stiffness is accepted as an important determinant of increased PP, thus providing predictive value for CV outcomes.32
In order to clarify the potential mechanisms of LVDD-PEF in view of epigenetic modifications, DNA methylation analysis was further performed. To our knowledge, it is the first study to simultaneously compare the DNA methylation difference in the early-stage of HF. Emerging evidence of epigenetic mechanisms is considered to be involved in the progression of HF, resulting in cardiac remodelling and accelerating cardiac dysfunction.33 Movassagh et al discovered the changes of gene expression by genome-wide maps of DNA methylation and histone-3 lysine-36 trimethylation in human end-stage hearts.34 Differential DNA methylation in the region of promoters of upregulated genes was shown. Xiao et al suggested that increased DNA methylation played a causative role in reducing cardiac contractility.11 Three differential DNA methylation genes including RGS10, CRACR2A and ACTG2 were identified as potential epigenetic markers for predicting LVDD-PEF in this study.
The regulator of G-protein signaling (RGS) proteins had been found to be engaged in diverse pathophysiological progress in CVDs, such as arrhythmias35 and ischemic injury.36 Recently, RGS10 was identified as a negative regulator of cardiac remodelling and its overexpression reversed the hypertrophy induced by aortic banding both in murine and human hearts;37 whereas hypomethylated alteration of RGS10 was observed in the region of the promoter for those presented end-stage HF.34 How RGS10 acts during the progress of the early-stage of HF still remains unclear. Accordingly, our observed finding in alteration of DNA methylation of RGS10 might be a result of enhanced expression of RGS10 protein, which needs further interpretation in subsequent studies. Besides, left ventricular abnormalities of in HFpEF might be compounded by microvascular dysfunction.30 Calcium release activated channel regulator 2A (CRACR2A) protein coded by CRACR2A is a modulator of calcium-release-activated calciumchannels which plays a role in endothelial cells. Previous evidence found that it was a large Rab GTPase and acted in intracellular signaling pathways. One of the important features of CRACR2A is GDP binding.38 Expression of mRNA was detected in cardiac microvasculature endothelial cell as well. A significant contribution to endothelial remodelling was observed through CRACR2A.38 The function of CRACR2A in CV system was still unclear since there were few studies focused in this field. Actin gamma smooth muscle 2 (ACTG2) was enriched the pathway of vascular smooth muscle contraction provided according to KEGG. Normally, actin proteins took part in the intracellular process, incorporating maintenance of cytoskeleton. ACTG2 was generally discovered in enteric tissue and abnormally expressed in different cancer types.39 However, the mechanisms underlying HF regulated by ACTG2 still remained unclear.
The strengths of our study were: firstly, our survey derived from large community-based samples, the results were therefore representative when extrapolating to the whole population; secondly, we evaluated potential risk factors, as well as epigenetic markers for predicting LVDD-PEF; finally, reliable echocardiographic parameters were available at baseline in our study to assess the diastolic function accurately.
Several limitations have to be interpreted in the present study: first, this is a cross-sectional study limited the actual relationship evaluation. Nevertheless, we will be able to provide more data at the 5 years’ follow-up visit since the Northern Shanghai Study is an on-going prospective study; additionally, mechanisms of these differentially methylated genes associated with effects of DNA methylation have not been validated on the development of LVDD-PEF.
This community-based study of senior residents revealed that female gender, BMI, greater PP and CIMT were significantly associated with LVDD-PEF. Differentially methylated genes such as RGS10, CRACR2A and ACTG2 were suggested to be further identified for predicting LVDD-PEF using in vitro and in vivo assays.
Data Sharing Statement
Individual deidentified participant data underlying the results reported in this study and other study-related documents will not be accessible since the Northern Shanghai Study is an on-going prospective community-based study. The follow-up study is still in the process and new enrolment is also conducted at the same time.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant no. 81872720) and Shanghai Municipal Commission of Health and Family Planning (grant no. 201840066) to Dr Lijuan Zhang. The National Natural Science Foundation of China (grant no. 81471402) to Jue Li and the National Natural Science Foundation of China (grant no. 81501203) to Jie Zhang. The abstract of this paper was presented at the Pulse of Asia 2019 Shanghai Conference name “Risk factors and epigenetic markers of left ventricular diastolic dysfunction with preserved ejection fraction in a community-based elderly Chinese population” as a poster presentation with interim findings. The poster’s abstract was published in “Poster Abstracts” in Pulse: DOI: 10.1159/000499586.
Author Contributions
All authors contributed to the conception and design, data analysis, drafting and critically revising the article, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Roger VL, Weston SA, Redfield MM, et al. Trends in Heart Failure Incidence and Survival in a Community-Based Population. JAMA. 2004;292(3):344–350. doi:10.1001/jama.292.3.344
2. Schocken DD, Benjamin EJ, Fonarow GC, et al. Prevention of heart failure: a scientific statement from the American Heart Association Councils on Epidemiology and Prevention, Clinical Cardiology, Cardiovascular Nursing, and High Blood Pressure Research; Quality of Care and Outcomes Research Interdisciplinary Working Group; and Functional Genomics and Translational Biology Interdisciplinary Working Group. Circulation. 2008;117:2544–2565. doi:10.1161/CIRCULATIONAHA.107.188965
3. Ponikowski P, Voors AA, Anker SD, et al. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: the Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC). Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur J Heart Fail. 2016;18:891–975. doi:10.1002/ejhf.592
4. Nagueh SF, Smiseth OA, Appleton CP, et al. Recommendations for the evaluation of left ventricular diastolic function by echocardiography: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. Eur Heart J Cardiovasc Imaging. 2016;17:1321–1360. doi:10.1093/ehjci/jew082
5. van Riet EE, Hoes AW, Wagenaar KP, Limburg A, Landman MA, Rutten FH. Epidemiology of heart failure: the prevalence of heart failure and ventricular dysfunction in older adults over time. A systematic review. Eur J Heart Fail. 2016;18:242–252. doi:10.1002/ejhf.483
6. Aljaroudi W, Alraies MC, Halley C, et al. Impact of progression of diastolic dysfunction on mortality in patients with normal ejection fraction. Circulation. 2012;125:782–788. doi:10.1161/CIRCULATIONAHA.111.066423
7. Kane GC, Karon BL, Mahoney DW, et al. Progression of left ventricular diastolic dysfunction and risk of heart failure. JAMA. 2011;306:856–863. doi:10.1001/jama.2011.1201
8. Yancy CW, Jessup M, Bozkurt B, et al. 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2013;62:e147–e239. doi:10.1016/j.jacc.2013.05.019
9. Berezin A. Epigenetics in heart failure phenotypes. BBA Clin. 2016;6:31–37. doi:10.1016/j.bbacli.2016.05.005
10. Napoli C, Grimaldi V, De Pascale MR, Sommese L, Infante T, Soricelli A. Novel epigenetic-based therapies useful in cardiovascular medicine. World J Cardiol. 2016;8:211–219. doi:10.4330/wjc.v8.i2.211
11. Xiao D, Dasgupta C, Chen M, et al. Inhibition of DNA methylation reverses norepinephrine-induced cardiac hypertrophy in rats. Cardiovasc Res. 2014;101:373–382. doi:10.1093/cvr/cvt264
12. Ji H, Xiong J, Yu S, et al. Northern Shanghai Study: cardiovascular risk and its associated factors in the Chinese elderly-a study protocol of a prospective study design. BMJ Open. 2017;7:e013880. doi:10.1136/bmjopen-2016-013880
13. Chobanian AV, Bakris GL, Black HR, et al. The seventh report of the joint national committee on prevention, detection, evaluation, and treatment of high blood pressure: the JNC 7 report. JAMA. 2003;289:2560–2572. doi:10.1001/jama.289.19.2560
14. Lang RM, Bierig M, Devereux RB, et al. Recommendations for chamber quantification: a report from the American Society of Echocardiography’s Guidelines and Standards Committee and the Chamber Quantification Writing Group, developed in conjunction with the European Association of Echocardiography, a branch of the European Society of Cardiology. J Am Soc Echocardiogr. 2005;18:1440–1463. doi:10.1016/j.echo.2005.10.005
15. Teichholz LE, Kreulen T, Herman MV, Gorlin R. Problems in echocardiographic volume determinations: echocardiographic-angiographic correlations in the presence of absence of asynergy. Am J Cardiol. 1976;37:7–11. doi:10.1016/0002-9149(76)90491-4
16. Zhang Y, Li Y, Liu M, Sheng CS, Huang QF, Wang JG. Cardiac structure and function in relation to cardiovascular risk factors in Chinese. BMC Cardiovasc Disord. 2012;12:86. doi:10.1186/1471-2261-12-86
17. Nagueh SF, Appleton CP, Gillebert TC, et al. Recommendations for the evaluation of left ventricular diastolic function by echocardiography. Eur J Echocardiogr. 2009;10:165–193. doi:10.1093/ejechocard/jep007
18. Paulus WJ, Tschope C, Sanderson JE, et al. How to diagnose diastolic heart failure: a consensus statement on the diagnosis of heart failure with normal left ventricular ejection fraction by the Heart Failure and Echocardiography Associations of the European Society of Cardiology. Eur Heart J. 2007;28:2539–2550. doi:10.1093/eurheartj/ehm037
19. Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi:10.1186/1471-2105-10-232
20. Abdi H. Bonferroni and Sidak corrections for multiple comparisons. In: Salkind NJ, editor. Encyclopedia of Measurement and Statistics. Thousand Oaks: Sage; 2007:103–107.
21. Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi:10.1093/nar/gkm882
22. Borlaug BA, Paulus WJ. Heart failure with preserved ejection fraction: pathophysiology, diagnosis, and treatment. Eur Heart J. 2011;32:670–679. doi:10.1093/eurheartj/ehq426
23. Sharma K, Kass DA. Heart failure with preserved ejection fraction: mechanisms, clinical features, and therapies. Circ Res. 2014;115:79–96. doi:10.1161/CIRCRESAHA.115.302922
24. Fischer M, Baessler A, Hense HW, et al. Prevalence of left ventricular diastolic dysfunction in the community. Results from a Doppler echocardiographic-based survey of a population sample. Eur Heart J. 2003;24:320–328. doi:10.1016/s0195-668x(02)00428-1
25. Goncalves A, Almeida PB, Lourenco P, Alvelos M, Betrencourt P, Azevedo A. Clinical significance of impaired relaxation pattern in middle-aged and elderly adults in the general population. Rev Port Cardiol. 2010;29:1799–1806.
26. Abhayaratna WP, Smith WT, Becker NG, Marwick TH, Jeffery IM, McGill DA. Prevalence of heart failure and systolic ventricular dysfunction in older Australians: the Canberra Heart Study. Med J Aust. 2006;184:151–154.
27. Mosterd A, Hoes AW, de Bruyne MC, et al. Prevalence of heart failure and left ventricular dysfunction in the general population; The Rotterdam Study. Eur Heart J. 1999;20:447–455.
28. Perkiomaki JS, Mottonen M, Lumme J, Kesaniemi YA, Ukkola O, Huikuri HV. Predictors of Development of Echocardiographic Left Ventricular Diastolic Dysfunction in the Subjects Aged 40 to 59 Years (from the Oulu Project Elucidating Risk of Atherosclerosis Study). Am J Cardiol. 2015;116:1374–1378. doi:10.1016/j.amjcard.2015.07.054
29. Cil H, Bulur S, Turker Y, et al. Impact of body mass index on left ventricular diastolic dysfunction. Echocardiography. 2012;29:647–651. doi:10.1111/j.1540-8175.2012.01688.x
30. Butler J, Fonarow GC, Zile MR, et al. Developing therapies for heart failure with preserved ejection fraction: current state and future directions. JACC Heart Fail. 2014;2:97–112. doi:10.1016/j.jchf.2013.10.006
31. Payne JR, James LE, Eleftheriou KI, et al. The association of left ventricular mass with blood pressure, cigarette smoking and alcohol consumption; data from the LARGE Heart study. Int J Cardiol. 2007;120:52–58. doi:10.1016/j.ijcard.2006.08.043
32. Laurent S, Alivon M, Beaussier H, Boutouyrie P. Aortic stiffness as a tissue biomarker for predicting future cardiovascular events in asymptomatic hypertensive subjects. Ann Med. 2012;44 Suppl 1:S93–S97. doi:10.3109/07853890.2011.653398
33. Di Salvo TG, Haldar SM. Epigenetic mechanisms in heart failure pathogenesis. Circ Heart Fail. 2014;7:850–863. doi:10.1161/CIRCHEARTFAILURE.114.001193
34. Movassagh M, Choy MK, Knowles DA, et al. Distinct epigenomic features in end-stage failing human hearts. Circulation. 2011;124:2411–2422. doi:10.1161/CIRCULATIONAHA.111.040071
35. Qin M, Huang H, Wang T, et al. Absence of Rgs5 prolongs cardiac repolarization and predisposes to ventricular tachyarrhythmia in mice. J Mol Cell Cardiol. 2012;53:880–890. doi:10.1016/j.yjmcc.2012.10.003
36. Parra S, Huang X, Charbeneau RA, et al. Conditional disruption of interactions between Galphai2 and regulator of G protein signaling (RGS) proteins protects the heart from ischemic injury. BMC Pharmacol Toxicol. 2014;15:29. doi:10.1186/2050-6511-15-29
37. Miao R, Lu Y, Xing X, et al. Regulator of G-protein signaling 10 negatively regulates cardiac remodeling by blocking mitogen-activated protein kinase-extracellular signal-regulated protein kinase 1/2 signaling. Hypertension. 2016;67:86–98. doi:10.1161/HYPERTENSIONAHA.115.05957
38. Wilson LA, McKeown L, Tumova S, Li J, Beech DJ. Expression of a long variant of CRACR2A that belongs to the Rab GTPase protein family in endothelial cells. Biochem Biophys Res Commun. 2015;456:398–402. doi:10.1016/j.bbrc.2014.11.095
39. Edfeldt K, Hellman P, Westin G, Stalberg P. A plausible role for actin gamma smooth muscle 2 (ACTG2) in small intestinal neuroendocrine tumorigenesis. BMC Endocr Disord. 2016;16:19. doi:10.1186/s12902-016-0100-3
40. Levey AS, Stevens LA, Schmid CH, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604–612. doi:10.7326/0003-4819-150-9-200905050-00006
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

