Back to Journals » Drug Design, Development and Therapy » Volume 13
Recent advances in the treatment of pathogenic infections using antibiotics and nano-drug delivery vehicles
Authors Giau VV , An SSA , Hulme J
Received 11 October 2018
Accepted for publication 30 November 2018
Published 18 January 2019 Volume 2019:13 Pages 327—343
DOI https://doi.org/10.2147/DDDT.S190577
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Tuo Deng
Vo Van Giau, Seong Soo A An, John Hulme
Department of Bionano Technology, Gachon Bionano Research Institute, Gachon University, Seongnam-si, Gyeonggi-do, South Korea
Abstract: The worldwide misuse of antibiotics and the subsequent rise of multidrug-resistant pathogenic bacteria have prompted a paradigm shift in the established view of antibiotic and bacterial–human relations. The clinical failures of conventional antibiotic therapies are associated with lengthy detection methods, poor penetration at infection sites, disruption of indigenous microflora and high potential for mutational resistance. One of the most promising strategies to improve the efficacy of antibiotics is to complex them with micro or nano delivery materials. Such materials/vehicles can shield antibiotics from enzyme deactivation, increasing the therapeutic effectiveness of the drug. Alternatively, drug-free nanomaterials that do not kill the pathogen but target virulent factors such as adhesins, toxins, or secretory systems can be used to minimize resistance and infection severity. The main objective of this review is to examine the potential of the aforementioned materials in the detection and treatment of antibiotic-resistant pathogenic organisms.
Keywords: antibiotics, resistance, polymer, chitosan, gold, recombinant, targeted, pathogen
Introduction
Antibiotics are compounds that either stop bacteria from growing or kill them outright, depending on their ability to inhibit critical bacterial cellular processes. Despite the profound success achieved by the use of broad-spectrum antibiotics in the treatment of complex bacterial diseases, new and recurrent bacterial infections continue to impose significant challenges on global health care.1–5 Conventional broad-spectrum therapies kill native as well as pathogenic bacteria disrupting the indigenous microflora increasing the risk of infection recurrence and systemic infiltration of the host.6–8 Recent studies suggest a number of pathogens such as Mycobacterium tuberculosis, Burkholderia pseudomallei, Staphylococcus aureus, and Listeria monocytogenes resist free form antibiotics by persisting in macrophages and erythrocytes.9,10 Some of these persistors remain dormant and can go undetected by standard culture methods.11 For the last fifty years identification of antibiotic resistant strains relied on selective culturing (blood cultures) and enzyme linked immune-sorbent assays (ELISA).12 Selective culturing for the diagnosis of pathogens takes from 18 to 48 hours.13,14 Unfortunately, these techniques are not economically feasible in low-resource settings where resistance rates exceed 80%.15,16 In 2012, the WHO published a list of critically important antimicrobials for human medicine (Table 1). Taken together, these challenges highlight the need for alternative antimicrobial detection and treatment strategies.
  | Table 1 WHO listing (third revision, 2012)16 of critically important antimicrobials for human medicine |
In the last few decades, many studies have focused on developing rapid detection and efficient drug delivery systems to meet the challenge of antibiotic resistance. These delivery vehicles can be nano or micron in size and composed of liposomes, dendrimers, peptides, polymer, and inorganic materials.17,18 Many of them are compatible with and have shown to enhance the sensitivities of conventional laboratory and field diagnostic tests by several orders of magnitude. The engineering of these vehicles is now so advanced that single bacteria can be routinely detected by point-of-care (POC) devices.19
In addition to detection, nano-vehicles are also used to treat bacterial infections. A drug or drugs can be loaded to the exterior or interior of the vehicle via chemical conjugation, adsorption, and encapsulation and then delivered to the infection site. Antibiotics delivered in this way shield the drug from the resistant mechanisms of the pathogen. Moreover, if the vehicle itself exhibits antimicrobial activity, the concentration of antibiotic can be reduced further. Other advantages of this combinatorial approach include longer drug retention times, improved pharmacokinetic profiles, and minimization of adverse effects to the host. These advances come at a time when the dissemination of metallo-β-lactamases among Gram-negative bacteria is growing dramatically and the resistance to carbapenem and β-lactam antibiotics exceeds 50% in many patient groups.20 As a result, several of these antibacterial vehicles are undergoing preclinical and clinical testing, while numerous others have recently been approved in the treatment of bacterial diseases21–30 by The US Food and Drug Administration such as silver and titanium nanoparticles (NPs) for antibacterial skin lotions and sunscreens that are in commercial use.
In this review, five areas where biocompatible nanomaterials can be used to combat antibiotic-resistant pathogenic bacteria are discussed. These areas include 1) detection of antibiotic-resistant pathogens, 2) targeted antibiotic treatments, 3) combinatorial antibiotic delivery, 4) NP vaccines, and 5) antibiotic repotentiation. Advances made in these areas bring us ever closer to real-time antibacterial therapies that can respond to changes in bacterial susceptibility, colonization resistance, infiltration of the host organism, and pathogenic virulence. In this review, we highlight the innovative materials currently being developed to detect and treat antibiotic-resistant pathogens.31–36
Pathogenic detection
The rapid identification of pathogenic bacteria in the early stages of therapy is crucial in preventing drug resistance. Among the myriad of molecular biology detection techniques available in modern day laboratories, those based on nucleic acid amplification technology such as whole genome sequencing, modern PCR, multiplex PCR (mPCR), quantitative PCR (qPCR), and droplet PCR are routinely used to distinguish resistant pathogenic strains. Real-time PCR (RT-PCR) or qPCR is capable of continuously monitoring changes in bacterial susceptibility.37 However, RT-PCR is currently too expensive to be employed in resource-limited settings where cost-effective qualitative tests dominate the field.38 In this regard, NPs can improve the sensitivity, accuracy, and reliability of qualitative tests, leading to better patient outcomes.
The first stage in identification involves sample preparation. This can take several days using gold standard techniques, which may be too long for critically ill patients who require administration of a specific antibiotic within 24 hours. Immunomagnetic separation is a rapid and highly facile extraction process that allows the researchers to concentrate microbial organisms from various clinical samples within hours.39 In the last decade, magnetic nanoparticles (MNPs) particularly superparamagnetic nanoparticles (SPMNPs) composed of magnetite (Fe3O4) or maghemite (γ-Fe2O3) are attracting a lot of attention due to their excellent magnetic properties, low cost, assay versatility, and higher capture efficiency.40 In addition to sample preparation, SPMNPs can be used directly or as part of multifunctional composite to improve the sensitivity (eg, Fe3O4-Ag, FeO4-Au, and FePt-Ag) of disposable optical and electrochemical immunoassays. For example, in 2017, Kearns et al combined lectin-functionalized silver-coated MNPs with optically active antibody-coated silver NPs to isolate and detect three bacterial pathogens including methicillin-resistant S. aureus (MRSA) in an Eppendorf tube.41 In 2018, Cowger et al developed a novel susceptibility test called protein-adsorbed NP-mediated matrix-assisted laser desorption–ionization mass spectrometry. The whole procedure took <50 minutes and was able to distinguish between wild-type and drug-resistant strains of bacteria.42 In addition, Wei et al reported a portable multiplexed bar-chart SpinChip (MB-SpinChip) integrated with NP-mediated magnetic aptasensors for the visual quantitative instrument-free detection of multiple pathogen. Using this MB-SpinChip, three major foodborne pathogens including Salmonella enterica, Escherichia coli, and L. monocytogenes were specifically quantified in apple juice with limits of detection of about 10 CFU/mL. Although significantly more expensive to construct than paper-based devices, MB-SpinChip may provide essential “real time” input during a potential pathogenic outbreak.43
Antibody-labeled SPMNPs are routinely used with standard and miniaturized MRI systems found in large hospitals and research laboratories. Diagnostic assays using only magnetic fields are simpler to use and better suited for the detection of bacteria in optically opaque media.44,45 In 2013, Chung et al used a magnetic-DNA probe in combination with a miniaturized nuclear magnetic resonance to detect the bacterial RNA of 13 species from patient specimens. The miniaturized micronuclear magnetic resonance system had a sample volume of 2 mL and capable of supporting rapid, high-throughput operations in POC settings.46 More recently, Park et al used a chip magnetic capture, culture, and detection assay for on-site detection of antigen-presenting cells exposed to S. enterica. The magnetic capture, culture, and detection achieved high sensitivity, detecting Salmonella-specific antibodies secreted from as little as 50 hybridoma cells/mL of blood. The assay took a minimum of 19 hours, 5 hours less than conventional tests and could be performed with unskilled labor.47 A schematic diagram demonstrating the capture and culturing stages of the assay is shown in Figure 1.
  | Figure 1 Checkerboard magnetic chip for cell capture. |
Using noninvasive imaging such as MRI to locate bacteria during the early stages of infection has the potential to reduce bacterial resistance and minimize organ damage to the host. Conventional MRI-based imaging has also been used to visualize S. aureus infections in endocarditis,48 osteomyelitis,49 and soft tissue infection models.50,51 Generally, these models detect inflammation rather than the causative agent of an infection. Although direct visualization of bacteria has been achieved using iron particle-labeled S. aureus,52 this technique is frequently limited by the lack of specificity. Ongoing work to create specific molecular imaging probes includes using iron NPs conjugated with IgG to detect intracellular pathogens, and particles coated with M. tuberculosis surface antibody for the specific detection of extrapulmonary mycobacterial infection53,54 have been reported.
For the past 3 decades, the optical and electrochemical properties of noble metal NPs, particularly AuNPs, have been extensively utilized by many fields of academic and industrial science.55,56 AuNPs exhibit a bright red maximum in the visible region of the optical spectrum; the maxima’s bandwidth and intensity can be tuned by varying the shape, composition, and the distance between particles.57 Aggregation of NPs induces interparticle surface plasmon coupling that results in a blue shift in the maxima. This colorimetric change has been utilized in numerous biological and NP immunoassays. It should be noted that the sensitivity for colorimetric sensors is low compared with fluorescence-based sensors, although the associated instrumentation is significantly cheaper. Moreover, many types of colorimetric sensors are designed to detect clinically relevant bacterial concentrations such as those found on human skin, although more sensitive methodologies are sometimes employed when the individual is suspected to be infected with a hypervirulent pathogen. For a comprehensive understanding of surface plasmon resonance in gold NPs, the review by Amendola et al is recommended.58
Pathogenic bacteria were first detected using AuNP aggregation by Elghanian et al in 1997.59 Since then, aggregation and dispersion of AuNPs have been widely explored for the detection of bacteria-specific DNAs, proteins, and live cells. In 2004, Storhoff et al used AuNPs to detect the mecA gene in MRSA genomic DNA samples.60 The approach was effective in discriminating MRSA from methicillin-sensitive S. aureus strains. More recently, Chan et al also used AuNPs for direct colorimetric PCR detection of MRSA in clinical specimens, a total of 72 clinical samples were tested.61 The performance was comparable with real-time PCR assays but at lower cost per reaction. Similar technological approaches have been applied in the detection of multidrug-resistant M. tuberculosis and hemorrhagic E. coli.62 Of particular interest is the application of these technologies in low-resource settings. In 2017, a working concept of an integrated paper microwell platform and an M. tuberculosis detection scheme based on AuNPs was demonstrated.63 Moreover, continued improvements in micropatterned paper devices combined with digitized fluidics have led to an increase in the availability of affordable, accurate POC tests. There are several up-to-date reviews regarding the detection of antibiotic-resistant strains of pathogenic bacteria using POC.64,65 Unfortunately, the potential of AuNPs is yet to be realized in the clinic, as many nanodiagnostic tests are stuck in preclinical trials with few US Food and Drug Administration tests currently available to the patient.
In addition to iron-based and gold NPs, quantum dots (QDs), upconverting nanoparticles (UCNPs), and graphene oxide (GO) have also been used to detect bacteria.66,67 Fluorescent QDs and UCNPs exhibit greater sensitivity and specificity than nonfluorescent nanomaterials as only the signal originating from the nanomaterial is detected. Graphene can be combined with QDs to work as a quencher (silencing the fluorescence signal of QDs when both are in contact).68 This property was utilized by Morales-Narváez et al in the fabrication of lateral flow biosensors.69 Their system consisted of two lines of QDs deposited on paper (test and control). The first line was capable of capturing some bacteria (by means of antibodies attached on QDs). After dispensing the sample solution on the QD lines, a solution of GO was added. In the absence of bacteria, GO quenched the fluorescence of the lines. In comparison with traditional lateral flow biosensors, this simple referencing method prevented the formation of false positives as a negative sample always turned off both lines. More recently, Jin et al utilized the dissociation of UCNPs and fluorescence recovery to detect E. coli in food and water samples, exhibiting a detection range of 5–106 CFU/mL and detection limit of 3 CFU/mL, respectively.70 The novel UCNP-based fluorescence resonance energy transfer (FRET) aptasensor could be used to detect a broad range of targets from whole cells to metal ions.
Targeted antibiotic therapies
The microenvironment of a bacterial infection site is rich in physiological and molecular cues that can be utilized to activate drug release or facilitate the binding of an NP or vehicle to bacteria. Passive targeting (natural barriers), physical–chemical gradients (surface charge and pH), and active targeting are three ways in which targeted drug delivery can be achieved.71,72 Passive targeting is possible due to the dysfunctional lymphatic drainage and elevated permeability that occurs at the infected site. This phenomenon can be exploited by nano and micron particles for targeted antibiotic delivery. In fact, the accumulation of both uncoated and PEGlyated liposomes at soft tissue infected by S. aureus has been shown to correlate closely with particle size.73,74
However, most infection sites are polymicrobial in nature.75 For example, pulmonary infections often consist of combinations of Gram-negative and Gram-positive anaerobic or aerobic bacteria, such as Klebsiella pneumoniae, E. coli, Pseudomonas aeruginosa, and S. aureus including MRSA.75,76 Many of these bacteria present a negative surface charge under physiological pH and can be targeted using common ligands such as cationic or lipopeptides.77 Cationic antimicrobial peptides and lipopeptides kill the microbes by interacting and disrupting bacterial cell membranes. Consequently, a range of vehicles incorporating natural and synthetic peptides have been developed for broad-spectrum applications.79 Moreover, lipopeptides show remarkable growth inhibition activity of both Gram-positive and Gram-negative bacteria and fungus. It should be noted that there is a potential for electrostatic interactions and subsequent binding of cationic peptides with other charge modifying substances in circulation. Studies have shown potentially toxic in vitro and in vivo effects with the use of cationic lipids and polymer, including but not limited to cell shrinking and vacuolization of human cell lines.80 Recently, Mitra et al showed that synthetic lipopeptides can exhibit good biocompatibility to different mammalian cell lines like HepG2, HeLa, and SiHa as well.81
Utilizing surface negative charge is one way to target the infection site; however, some pathogens can negate this approach by secreting lactic and acetic acid into the microenvironment.82 To overcome this problem, several groups have developed surface charge switching polymer micelles and NPs that target the negatively charged cell wall of MRSA at low pH.83,84 Recently, a zwitterionic pH-sensitive polymer, poly (N′-citraconyl-2-(3-aminopropyl N, N dimethylammonium) ethyl methacrylate) or P(CitAPDMAEMA), was used to inhibit the growth of E. coli and S. aureus under acidic conditions.85 The polymer remained zwitterionic at physiological pH and exhibited low hemotoxicity and good biocompatibility. Surface charge switching micelles and NP have been synthesized containing pH-responsive poly-l-histidine,84 beta-amino ester,86 and numerous chemical functionalities.87 In addition, polymers containing acid-degradable acetal/ketal functionalities have also shown promising results for pH-responsive drug delivery systems for anticancer drugs.88 With this in mind, Kalhapure et al recently developed a pH-responsive solid lipid NP with an acetal linkage to deliver vancomycin and inhibit the growth of methicillin-sensitive S. aureus and MRSA. The in vivo study showed that the amount of MRSA remaining in the skin of encapsulated vancomycin-treated mice was ~22-fold lower than those treated with the free form of the drug.89
In addition to MRSA, many bacteria such as Helicobacter pylori, Listeria, Salmonella, Campylobacter rectus, E. coli, and Shigella flexneri are found in varying acidic conditions within the body (stomach [pH 1.0–2.0], vagina [pH 4.0–5.0], skin [pH 4.0–5.5], intestines [pH 5.0–8.0] and bladder [pH 4.5–8.0]).84 The stomach mucosa is particularly vulnerable to the negative spiral bacterium H. pylori, which is etiologically associated with the development of gastritis, gastric ulcers, and associated carcinomas. The best results regarding the eradication of H. pylori have been achieved using a variety of antibiotic-loaded chitosan (CS) microparticles and NPs.90–93 Despite these successes, the physical–chemical requirements that determine NP assembly on different nanostructured H. pylori surfaces are still not resolved.94 Recent work by Westmeier et al attempted to address this issue by studying the behavior of a library of model NPs, varying in size, surface functionalization, shape, and material on H. pylori.95 The study showed that NP–bacteria assembly did not require specific functionalization or negative surface charge and that assembly in interfering medium was significantly influenced by the impact of low pH on the bacterial surface. Moreover, assembly of nonbactericidal silica NPs (25% surface coverage) was shown to attenuate the pathogenesis of H. pylori by probable inhibition of the type IV secretion system (Figure 2).
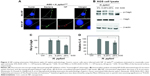  | Figure 2 NP-coating attenuates Helicobacter pylori (H. pylori) pathobiology. Gastric cancer cells were infected with NP -H. pyloriGFP complexes estimated to maximally cover approximately 25% (Si30–25%: 1 × 108 bacteria, 600 μg mL−1 Si30; 10 min PBS) or 0.25% (Si30–0.25%: 1 × 108 bacteria, 60 μg mL−1 Si30, 10 min PBS) of the bacterial surface, respectively. |
NPs have shown promise in treating stomach bacterial infections, but a significant challenge has been to develop antibacterial NPs that are suitable for the various pH gradients found in the gastrointestinal tract. One such material is the triblock copolymer poly(ethylene glycol)-block-poly (aminolated glycidyl methacrylate)-block-poly(2-(diisopropyl amino) ethyl methacrylate) (PEG-b-PAGA-b-PDPA), which was recently used as a pH-responsive micelle to target metastatic breast cancer.96
In addition to negative charge and pH gradients, bacteria secrete many virulence factors including toxins, fibronectin proteins, collagen adhesins, and various enzymes (phosphatase, phospholipase, and lipase).97 Thus, a simple way to target the infection site is to dope the surface of the antibiotic delivery vehicle with a complementary substrate or antibody. As the enzyme metabolizes the substrate, it perforates the vehicle releasing the antibiotic near the infection site. In the last decade, various lipase or phosphatase-sensitive polymeric triple-layered nanogels have been used for the synchronized delivery of antibiotics at infection sites.98,99 Lipases can also be used to shield AMPs from nonspecific interactions with the host. For example, the lipase substrate poly(caprolactone-b-ethylene glycol) (PCL-b-PEG) was recently used to mask the dendritic polycation G-2. The hybrid dendrimer successfully killed >99.9% of inoculated bacterial cells at ≤8 μg/mL, exhibiting good colloidal stability in the presence of serum and insignificant hemolytic toxicity even at ≥2,048 μg/mL.100 The design of these hybrid vehicles is now so refined that it is possible to treat a specific pathogenic strain. For example, Yang et al recently reported a unique intracellular antibiotic delivery NP, in which mesoporous silica NPs loaded with gentamicin were coated with (bacterial toxin)-responsive lipid bilayer surface shell and bacteria-targeting peptide ubiquicidin (UBI29-41). The inhibition of S. aureus growth in both in vitro and in vivo of planktonic and intracellular infection, as well as a downregulation of inflammation-related gene expression in infected preosteoblasts or macrophages was observed.101
If the obligate or facultative bacterium escapes the infection site, the risk of infection recurrence increases. It is now accepted that bacteria escape by hiding (at least survive) in osteoblasts, erythrocytes, and phagocytic cells.9 These pathogens disrupt the microbicidal mechanisms of phagocytic cells by inhibiting the fusion of lysosomes with phagosomes prolonging their survival.102 Recent studies suggest that several pathogenic bacteria such as M. tuberculosis, B. pseudomallei, S. aureus, and L. monocytogenes are capable of this.10 Moreover, pathogens often use the cytoplasm of mammalian cells to exchange virulent and resistance genes.103 One of the key mechanisms by which microbe’s requestor macrophages is by subverting the body’s most prolific nuclear receptors, the vitamin D receptor.104 The impact of this subversion on melatonin homeostasis and the subsequent increase in intracellular infection are well known.105 Unfortunately, the development of vitamin D receptor agonists such as olmesartan medoxomil has yet to be fully realized.106
Once a macrophage becomes infected, the next step is to target the intracellular pathogen. As set out by Xiong et al,20 the optimal activity of a given antibiotic against any bacterial-infected cell depends on the concentration of the active drug within the subcellular compartments, which is dependent on the drugs cellular retention, subcellular distribution, and activity. One way to achieve this is to deliver the antibiotic to the cell or infection site via a biodegradable vehicle. Over the past 2 decades, several biodegradable delivery systems composed of natural or synthetic polymers such as poly(lactide-co-glycolide) (PLGA),18,107,108 poly(lactide) (PLA),109 albumin, and CS78 have demonstrated the potential of this approach. The major sites of intracellular infection are the mononuclear phagocytic system tissues found in the liver and spleen. This natural barrier brings together infected cells and antibiotic delivery vehicles amplifying the therapeutic effect of the antibiotic. This amplification is often reported when the mononuclear phagocytic system is the primary foci of infection in an experimental model. For example, ampicillin-loaded polyisohexylcyanoacrylate NPs demonstrated a therapeutic efficiency >2 orders better than the free form of the antibiotic in vivo.110 Similar results have been obtained using gentamicin-loaded nanoplexes based on block copolymers of poly(ethylene oxide-b-sodium acrylate) and poly(ethylene oxide-b-sodium methacrylate).111
Outside the liver and spleen, bacterial-infected macrophages are also found in the pulmonary cavities, eliminating these bacteria (active or dormant) remains one of the toughest challenges that developmental drug delivery systems currently face.112 For example, current treatment of pulmonary tuberculosis (TB) infection involves prolonged oral administration of high systemic doses of combined frontline antibiotics like rifampicin (RFP), isoniazid, pyrazinamide, and ethambutol over a 6- to 8-month period.113 M. tuberculosis can survive in alveolar macrophages by inhibiting phagosome maturation and phagosome–lysosome fusion. Delivery of RFP via PLGA microspheres to alveolar macrophages resulted in a 19-fold higher concentration of RFP in cells infected with TB compared with the administration of the free drug in solution.114 Uptake of the antibiotic by the infected alveolar macrophages was dependent on the size and concentration of vehicles used and the population of macrophages encountered at the infection site. Using PLGA microspheres to extend the release of the drug to the primary site of infection is a promising treatment for TB as it can achieve the desired therapeutic effect in a shorter period.115 Moreover, treatment time can be further reduced if the RFP-loaded PLGA microparticles are administered via intratracheal insufflation. When TB-infected guinea pigs were treated using the said procedure, their lungs showed significantly increased drug levels compared with intravenous and oral delivery of RFP.116
The rapid development of targeted therapies for TB has benefited from the unique surfaces of mycobacteria, making the targeting of the infection site easier. However, differentiating between two Gram species is more difficult.117 Yet, there are many common ligands (antibiotics included) that can be used to specifically target Gram-positive or Gram-negative pathogens.118 In the last 2 decades, various antibiotics such as vancomycin and amoxicillin have been conjugated to the surface of gold NPs,119,120 iron oxide NPs,121,122 porous silica NPs,123 and surface dendrimers, resulting in the preferential binding of Gram-positive bacteria. Moreover, other small molecules such as lectins, single domain antibodies, and aptamers124 can be conjugated to the surface of the delivery vehicle to target Gram-negative pathogens. For example, mannose-specific or fucose-specific lectins showed enhanced binding affinity to the carbohydrate receptors on H. pylori, while bacterium-based aptamer selection techniques have been utilized to target NPs to Salmonella typhimurium. In addition to these techniques, other groups have utilized cell–pathogen adhesion mechanisms to target opportunistic pathogens.125 Recently, Angsantikul et al utilized clarithromycin (CLR) loaded PLGA NPs (100 nm) coated with gastric epithelial cell membranes AGS-NP (CLR) to treat H. pylori infection in mice. After 6 days, the bacterial burden in a gram of stomach mice tissue treated with AGS-NP (CLR) was ~3.08 orders of magnitude lower than control (Figure 3).126
  | Figure 3 In vivo anti-Helicobacter pylori therapeutic efficacy of AGS-NP (CLR). |
Another pathogen that resides within human macrophages is MRSA. Studies examining infected alveolar macrophages found that PEGylated and non-PEGylated liposomes loaded with vancomycin were capable of significant improvements in MRSA clearance.127 Other types of delivery vehicles such as solid lipid NPs and PLGA loaded with vancomycin have shown similar improvements in MRSA clearance rates as well.128 More recently, Pei et al combined the pH-sensitive properties of CS with PEGylated-PLGA to form a hybrid vehicle namely PpZEV. PpZEV NPs exhibited increased vancomycin release at acidic pH and significantly higher levels of uptake and MRSA clearance in infected macrophages when compared with free vancomycin and unmodified PLGA-NPs.129 Macrophage uptake of antimicrobials can be further enhanced by attaching various ligands including O-stearoyl amylopectin, phosphatidylcholine, maleylated bovine serum albumin, and mannose onto the NP surface.130A summary of the various types of nanovehicles used to improve the efficacy of antibiotics and the treatment of bacterial infections is shown in Table 2.
Increasing microbial susceptibility: combinatorial applications
The accumulated misuse of free-form antibiotics has led to the emergence of multidrug-resistant MDR strains. The accepted definition of MDR is coresistance to three or more classes of antimicrobial drugs.131 A serious concern is Enterobacteriaceae (avian E. coli), which is known to carry the New Delhi metallo-beta-lactamase gene (NDM-1), as well as genes that confer resistance to copper sulfate and quaternary ammonium disinfectants.132 At present, most antibiotic-resistant mechanisms do not apply to NPs whose mode of action is direct contact with the cell wall. Thus, combining antibiotics with nanomaterials should reduce bacterial resistance and improve the pharmacokinetic profile of the drug.
Liposomes composed of phospholipids and cholesterols have been used to deliver antibacterial drugs for several decades.133 As a drug delivery system, liposomes offer several advantages. Using mice and guinea pig TB-infected models, Deol et al showed that RMP, INH, and PZA-encapsulated liposomes were significantly more effective in reducing the recovery times than their free drug counterparts.134 Due to the physical and chemical instability of liposomes, many researchers have employed niosomes to maintain the physical–chemical properties of antibiotic combinations.135 Niosomal delivery allows for the delivery of combinatorial antibiotics that are too toxic in their free form. For example, drug compounds such as bismuth derivatives and gallium desferrioxamine interfere with iron absorption and reduction of alginate, LPS, and the secretion of adhesion factors of P. aeruginosa.136 In a recent study by Mahdiun et al, the combinatorial effect of tobramycin and bismuth ethanedithiol-loaded niosomes (Nio-TOB) on the quorum sensing and biofilm production of P. aeruginosa was demonstrated. The minimum inhibitory concentration (MIC) of TOB and Nio-TOB for most strains was 16.64 and 1.3 mg/mL, respectively.137 P. aeruginosa uses the same uptake mechanisms for both gallium and iron.138 Thus, targeting bacterial iron metabolism can arrest P. aeruginosa virulence. Simple gallium siderophore complexes such as gallium citrate have shown good antibacterial activity (1–5 mg/mL MIC) against P. aeruginosa and low drug resistance in vivo. In addition, gallium protoporphyrin IX has shown 100- to 1,000-fold increase in potency against various MRSA strains compared with gallium nitrate.139 However, relatively low aqueous solubility, moderate toxicity, and low synthetic yields of Ga-protoporphyrin IX may have dampened enthusiasm for further development. Antibiotics can also be conjugated with bacterial siderophores increasing their membrane permeability, which in turn increases their antimicrobial activity. In 2017, Oh et al reported on a novel siderophore-conjugated cephalosporin, LCB10-0200. In accordance with Clinical and Laboratory Standards Institute methods, LCB10-0200 showed potent antibacterial activity against P. aeruginosa clinical isolates, including β-lactamase producing strains. Moreover, LCB10-0200 was more effective than ceftazidime in treating systemic, respiratory tract, urinary tract, and thigh infections caused by antibiotic susceptible and resistant strains of P. aeruginosa in mouse models.140
Until recently, first-line therapies for H. pylori eradication consist of proton pump inhibitors or ranitidine bismuth citrate and CLR, plus amoxicillin or metronidazole.141,142 Proton pump inhibitors and amoxicillin are known to increase the risk of secondary infections such as Clostridium difficile, while combinations of metronidazole are frequently used to treat C. difficile disease. Moreover, CLR resistance is now at a level where continued use of mainstream CLR-based triple therapy is not recommended.143,144 The clinical lifetime of CLR may have been prolonged if it had been encapsulated into a mucoadhesive particle. As early as 2008, several groups had tested various encapsulated triple combinations using NPs composed of CS, lectin-conjugated gliadin, and PGLA. Ramteke et al used lectin-conjugated NPs to simultaneously release amoxicillin, omeprazole, and CLR, resulting in a 94.83% eradication rate of H. pylori from infected rats.145
As >40% of the global population harbor H. pylori, a new treatment strategy is urgently needed.146 Fortunately, other types of eradication therapies such as liposomal linolenic acid (LipoLLA) have shown great potential in eradicating H. pylori. Unlike amoxicillin, LipoLLA kills both spiral and coccoid forms of the pathogen. However, a few studies suggested that certain carriers may cause adverse health effects by directly eliminating the gastrointestinal microbiota or by alternating their functions. For example, silver NPs could damage the gut microbiota even though silver is generally considered to have low toxicity. In a recent paper by Li et al, the gut microbiota of H. pylori-infected mice exhibited minor alterations in bacteria populations after LipoLLA therapy with a notable boost in the relative abundance of Proteobacteria and Firmicutes along with a decrease in Verrucomicrobia and Bacteroidetes.147 On the contrary, conventional triple therapy led to the growth of Enterobacteriaceae, Enterococcaceae, and Clostridiaceae species in the mice.
It is becoming increasingly obvious that the composition of the gut microbiome must be taken into consideration during the development and administration of future antibacterial treatments. Sparing the gut microbiome using narrow-spectrum antibiotics (fidaxomicin) has been shown to correlate with a reduction in C. difficile recurrence compared with broad-spectrum antibiotics.148 However, TB resistance is increasing although narrow spectrum drugs are the mainstay of current treatments. Moreover the oral-pulmonary microbiota is too small to act as a significant reservoir of antibiotic resistance, which suggests the administration of free form RFP, its rapid delocalization, and resulting gut metabolites have contributed significantly to an increase in its resistance.149
Vaccination vehicles (toxins and antigens)
As well as killing bacteria, the large surface area of NPs can be used to capture bacterial toxins. Many pore-forming toxins (PFTs) such as listeriolysin O (LLO) L. monocytogenes and Aerolysin from Aeromonas hydrophila promote deacetylation and dephosphorylation of histones, resulting in subsets of immune genes, such as CXCL2, MKP2, or IFIT3, becoming repressed thus predisposing the host to secondary infections. A radical but simple solution to accelerate the removal of active toxins from the host is to use erythrocyte membrane-coated polymer NPs or nanosponges.150 The membrane of red blood cells (RBCs) serves as perfect stealth coating with high biocompatibility and immune evasion ability.151–153 Using the extrusion method, various nanomaterials such as PGLA, melatonin, inorganic NPs, gold nanocages, and QDs have been coated with RBC membranes. In all cases, the in vivo circulatory half-life of RBC-coated nanomaterials was superior to materials coated with PEG. In 2017, Zhang et al prepared RBC membrane-coated vancomycin nanogels using the cell membrane-templated polymerization method for the treatment of MRSA infections.154 The redox-responsive hydrogel acted as the delivery vehicle for vancomycin, while the RBC membrane absorbed and neutralized the pore-forming toxin secreted by the bacteria. Compared with free antibiotics and nonresponsive nanogels, RBC-coated nanogels exhibited remarkable antibacterial activity in vitro. Recently, Chen et al developed a broad-spectrum nanosponge by coating 100 nm PLGA NPs with human RBC membranes.155 When tested with PFTs, melittin, αhemolysin, LLO (L. monocytogenes), and streptolysin O (group A Streptococcus), the hRBC nanosponges completely inhibited toxin-induced hemolysis in a concentration-dependent manner. The inhibitory human nanosponge concentration (IC100) value for the four PFTs tested was 7.5, 16, 0.63, and 1.0 μg/mL, respectively. The investigation showed that nanosponge sequestered toxins were less toxic not only to cells but also to live animals. Figure 4 shows a schematic structure of human nanosponge and its mechanism of neutralizing PFTs. In addition to α-hemolysin, MRSA also secretes other membrane-damaging toxins, including gamma-toxin, leucocidins, and phantom–valentine leucocidin. Such toxins can also be impregnated onto the surface of RBC-coated NPs. In 2017, Wei et al demonstrated a multitoxin laden nanotoxoid formulation using the aforementioned cell membrane-cloaked NPs.156 The formulation was nonhemolytic and showed no observable toxicity upon subcutaneous administration in mice. Immunization with the nanoformulation induced germinal center formation in the lymph node and increased antibody titers against three staphylococcal virulence factors. The vaccine was shown to be effective in reducing skin lesion formation and bacteria count in mouse model.
  | Figure 4 Structure of hNS. |
The unique capabilities of RBC membrane-coated NPs to detain toxins and induce an immunological response are well known.157–160 However, Gram-specific and mycobacterial outer membrane vehicles (OMVs) 50–250 nm can also be coated onto NPs. Given their small size, proinflammatory pathogen-associated molecule patterns, suitability for antigen-presenting cells uptake, and efficient lymph node drainage, OMVs are well suited to bacterial vaccine development. By marrying the virtues of synthetic NPs and OMVs, E. coli OMV-coated 30 nm gold NPs have been shown to induce fast dendritic cell activation and maturation in the lymph nodes of mice. The hybrid vehicles generated higher antibody titers with stronger avidity than those elicited by OMVs alone.161 In addition, the vaccinated mice showed elevated levels of IL-17 and interferon-gamma. The adjuvant activity of gold NPs has been used extensively in the development of antibacterial vaccines including L. monocytogenes, Yersinia pestis, P. aeruginosa, and Streptococcus pneumoniae.31 Multishell gold NPs have the potential to deliver bacterial antigens either through encapsulation or surface association. However, such investigations are usually conducted using cheaper biodegradable polymer NPs, such as PLGA NPs. For example, PLGA NP-encapsulated ovalbumin induced a higher and sustained antibody response than liposomes of similar size. It was also noted that the PLGA vaccine elicited a protective immunity against listeria infection with the highest bacterial clearance compared with liposomes. Antigen-coated polymer NPs have been shown to induce a strong cytotoxic CD8+ T cell response, whereas antigen encapsulation preferentially activates CD4+ T cells. However, several studies have shown that particle-encapsulated antigens induce CD8+ T cell as well, clearly further investigations on cellular processing of antigens is needed.162,163
To obtain protective immunity, adjuvants that augment the immunostimulating activity are required. Adjuvants can be formulated in the same way as antigens by encapsulation, incorporation, or conjugation with a polymer NP. Akin to bacteria, adjuvants that activate pattern recognition receptors (PRRs) can be added to the surface of antigen-loaded NPs. There are three major families of PRRs: Toll-like receptors (TLRs), retinoic acid-inducible gene I-like receptors, and nucleotide-binding oligomerization domain-like receptors. The cooperation of multiple PRRs has been shown to contribute to host defense against bacteria. Critical to the control of S. typhimurium is the synergy between TLR2 and TLR4, whereas the cooperation between active TLR2 and TLR9 plays an important role in controlling TB infection.164,165
As previously highlighted, antibiotic administration via pulmonary insufflation has proved very effective in the treatment of TB. Insufflation was employed by Lu et al to deliver Ag85B antigen and trehalose-6-6-dibehenate-loaded PLGA NPs to TB-infected guinea pigs, resulting in anti-TB protection, albeit inferior to BCG vaccination.166 A single toxin/antigen vaccine is often inadequate against MRSA or TB infection consequently researchers frequently adopt a triantigen approach. For example, three mycobacterial antigens, MPT-83, MPT-64, and Ag85B, were loaded into PLGA NPs by Cai et al and administered via intramuscular injection to mice. After vaccination, elevated cellular and humoral responses against all three antigens were observed. In particular, a single dose of the triantigenic formulation was comparable with the BCG vaccine.167
Antibiotic re-potentiation and gene editing
NP coatings (Ag, Au, ZnO, CuO, TiO2, etc) have been used to inhibit the growth of bacterial films on various surfaces (implants, dental materials, and wound dressings) for decades.168 Toxicity issues surrounding implant failure are beyond the scope of this review as are the coselective properties of ZnO, CuO, and TiO2 in animal feeds.13,23 In 2017, several in-depth reviews discussed the antibacterial mechanisms of free form inorganic NPs.23,34 More recently, work by Kadiyala et al showed that ZnO nanoparticles (ZnO-NPs) kill MRSA via ROS-independent mechanisms involving multiple metabolic pathways.169 Many reports suggest that these inorganic nanomaterials are toxic to specific cell lines and organs.170–173 However, when used at very low concentrations with antibiotics to treat infections they are not.174 A recent study by Panáček et al demonstrated a strong synergistic effect of antibiotics combined with AgNPs at very low concentrations demonstrating no cytotoxic effect on mammalian cells. Moreover, restoration of the susceptibility of a resistant E. coli strain to ampicillin was observed when ampicillin was combined with AgNPs. In addition, the reduced impact of fewer NPs on gut DNAases using the synergistic approach makes the uptake of resistant plasmids and transposons by live bacteria in humans unlikely.4
Much remains to be learnt about the exogenous and endogenous factors affecting antibiotic resistance. Changes in cell physiology can provide insight into the bacterial mechanisms contributing to antibiotic tolerance. Sugar potentiation of β-lactam antibiotics is now well established with arabinose, glucose, mannitol, ribose, and lactose capable of increasing the sensitivity of infectious strains of E. coli to carbenicillin and cefuroxime by several orders of magnitude.175,176 In addition to primary metabolite potentiation, nontoxic (human host) diarylamidines such as pentamidine have displayed synergy with antibiotics used to treat Gram-positive bacteria, yielding effective combinations in vitro to colistin-resistant bacteria.177,178 In addition, repotentiation of secondary metabolites (antibiotics) using metabolizable sugars and a diarylamidine was recently used to kill E. coli O157:H7 via a ROS mechanism while repressing Shiga toxin production.179 More recently, Milton et al180 presented a new urea-containing class of 2-aminoimidazole-based adjuvants that potentiated colistin activity against colistin-sensitive Acinetobacter baumannii. Lead compounds enabled 1,000-fold reduction in the MIC of colistin in vitro. It could be argued that there are many naturally occurring antibacterial polymers (CS) that can repotentiate last line antibiotics just as well. For example, recent work by Jamil et al synergistically enhanced the bactericidal activity of β-lactam antibiotic cefotaxime with antimicrobial biopolymer CS, rendering it more effective against biofilm producing MDR pathogens.181 Figure 5 shows the superior antibiofilm activity of cefotaxime-loaded CS NPs compared with CS NPs. Finally, interfering RNAi has been shown to repotentiate current antibiotics by targeting resistance genes. Recently, Meng et al used lipid nanocarrier to deliver antisense oligonucleotides targeted to the mecA gene, which is known to play a role in β-lactam resistance. Upon testing, they found it was possible to restore MRSA susceptibility to oxacillin in vitro and in vivo.182
  | Figure 5 CFU assay. |
In the event that these combinatorial approaches fail to arrest antibiotic resistance in animal models, the last line of defense is gene editing. Using a combination of clustered regularly interspaced, short palindromic repeats (CRISPR) and Cas9 protein, this accurate method of gene editing was recently applied as an anti-MRSA strategy in a study by Bikard et al.183 Using a delivery system known as a phagemid, it was possible to deliver the gene editing machinery to the MRSA resulting in a 104-fold reduction in the number of viable colonies in vitro. However, challenges arise due to the wildly varying size and structure of different phagemids; moreover, delivery of the CRISPR–Cas9 package to intracellular microbes could mitigate off-target effects at both host cell and bacterial level. Current delivery strategies involve in capsulation of the phagemid with biological inert polymers, lipids, silica, and DNA target.184,185 To the best of our knowledge, there are no reports of RBC membranes being used to deliver CRISPR/Cas9 machinery to antibiotic-resistant pathogens.
Conclusion
In the last decade, advances in bionanoengineering have contributed to the development of highly sensitive bacterial detection technologies and sensors. Major efforts in refining the interior and surface chemistry of organic and inorganic drug carriers such as CS, PLGA, and gold have demonstrated excellent outcomes via the detection treatment and prevention of bacterial diseases, by specific targeting of pathogenic bacteria, combinatorial delivery of antibiotics, and effective antibacterial vaccination strategies.
It is expected that advances in microbiology, digital microfluidics, and bionanoengineering will continue to bring improvements regarding the rapid isolation and detection of infectious pathogenic bacterial strains, improvements in cost-effective drug delivery systems, as well as tailored effective therapies in developed countries.
Unfortunately, the advantages of improved biodistribution and therapeutic outcomes using targeted nanomaterials have yet to be realized in developing countries where the need to combat antibiotic resistance is felt the strongest. Thus, the application of these advances will require communication and collaboration between scientists and clinicians from many different countries and fields of research.
Acknowledgment
This research was supported by the Gachon University Research Fund GCU-2016-0186.
Disclosure
The authors report no conflicts of interest in this work.
References
D’Costa VM, King CE, Kalan L, et al. Antibiotic resistance is ancient. Nature. 2011;477(7365):457–461. | ||
Martinez JL. General principles of antibiotic resistance in bacteria. Drug Discov Today Technol. 2014;11:33–39. | ||
McFall-Ngai M, Hadfield MG, Bosch TC, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A. 2013;110(9):3229–3236. | ||
van Schaik W. The human gut resistome. Philos Trans R Soc Lond B Biol Sci. 2015;370(1670):20140087. | ||
Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74(3):417–433. | ||
Potgieter M, Bester J, Kell DB, Pretorius E. The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol Rev. 2015;39(4):567–591. | ||
Najar MS, Saldanha CL, Banday KA. Approach to urinary tract infections. Indian J Nephrol. 2009;19(4):129–139. | ||
Kell D, Potgieter M, Pretorius E. Individuality, phenotypic differentiation, dormancy and “persistence” in culturable bacterial systems: commonalities shared by environmental, laboratory, and clinical microbiology. F1000Res. 2015;4:179. | ||
Pinto D, São-José C, Santos MA, Chambel L. Characterization of two resuscitation promoting factors of Listeria monocytogenes. Microbiology. 2013;159(Pt 7):1390–1401. | ||
Lungu B, Ricke SC, Johnson MG. Growth, survival, proliferation and pathogenesis of Listeria monocytogenes under low oxygen or anaerobic conditions: a review. Anaerobe. 2009;15(1–2):7–17. | ||
Audoly G, Fenollar F, Lagier JC, Lepidi H, Raoult D. Deglycosylation of Tropheryma whipplei biofilm and discrepancies between diagnostic results during Whipple’s disease progression. Sci Rep. 2016;6(1):23883. | ||
Kim ES, Kim HB, Kim G, et al; Korea INfectious Diseases (KIND) study group. Clinical and epidemiological factors associated with methicillin resistance in community-onset invasive Staphylococcus aureus infections: prospective multicenter cross-sectional study in Korea. PLoS One. 2014;9(12):e114127. | ||
Buchan BW, Ledeboer NA. Emerging technologies for the clinical microbiology laboratory. Clin Microbiol Rev. 2014;27(4):783–822. | ||
Cohen J, Vincent JL, Adhikari NK, et al. Sepsis: a roadmap for future research. Lancet Infect Dis. 2015;15(5):581–614. | ||
Lafleur LK, Bishop JD, Heiniger EK, et al. A rapid, instrument-free, sample-to-result nucleic acid amplification test. Lab Chip. 2016;16(19):3777–3787. | ||
Doyle MP, Loneragan GH, Scott HM, Singer RS. Antimicrobial resistance: challenges and perspectives. Compr Rev Food Sci Food Saf. 2013;12(2):234–248. | ||
Winnicka K, Wroblewska M, Wieczorek P, Sacha PT, Tryniszewska EA. The effect of PAMAM dendrimers on the antibacterial activity of antibiotics with different water solubility. Molecules. 2013;18(7):8607–8617. | ||
Gao W, Thamphiwatana S, Angsantikul P, Zhang L. Nanoparticle approaches against bacterial infections. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2014;6(6):532–547. | ||
Hulme J. Recent advances in the detection of methicillin resistant Staphylococcus aureus (MRSA). BioChip J. 2017;11(2):89–100. | ||
Xiong MH, Bao Y, Yang XZ, Zhu YH, Wang J. Delivery of antibiotics with polymeric particles. Adv Drug Deliv Rev. 2014;78:63–76. | ||
Couvreur P. Nanoparticles in drug delivery: past, present and future. Adv Drug Deliv Rev. 2013;65(1):21–23. | ||
Abed N, Couvreur P. Nanocarriers for antibiotics: a promising solution to treat intracellular bacterial infections. Int J Antimicrob Agents. 2014;43(6):485–496. | ||
Wang L, Hu C, Shao L. The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int J Nanomedicine. 2017;12:1227–1249. | ||
Pelgrift RY, Friedman AJ. Nanotechnology as a therapeutic tool to combat microbial resistance. Adv Drug Deliv Rev. 2013;65(13–14):1803–1815. | ||
Rai M, Ingle AP, Pandit R, et al. Broadening the spectrum of small-molecule antibacterials by metallic nanoparticles to overcome microbial resistance. Int J Pharm. 2017;532(1):139–148. | ||
Torres-Sangiao E, Holban A, Gestal MC. Advanced nanobiomaterials: vaccines, diagnosis and treatment of infectious diseases. Molecules. 2016;21(7):867. | ||
Carabineiro SAC. Applications of gold nanoparticles in nanomedicine: recent advances in vaccines. Molecules. 2017;22(5):E857. | ||
Rizvi SMD, Hussain T, Ahmed ABF, et al. Gold nanoparticles: a plausible tool to combat neurological bacterial infections in humans. Biomed Pharmacother. 2018;107:7–18. | ||
Assolini JP, Concato VM, Gonçalves MD, et al. Nanomedicine advances in toxoplasmosis: diagnostic, treatment, and vaccine applications. Parasitol Res. 2017;116(6):1603–1615. | ||
Hemeg HA. Nanomaterials for alternative antibacterial therapy. Int J Nanomedicine. 2017;12:8211–8225. | ||
Baptista PV, McCusker MP, Carvalho A, et al. Nano-strategies to fight multidrug resistant bacteria – “A Battle of the Titans”. Front Microbiol. 2018;9:1441. | ||
Yang K, Han Q, Chen B, et al. Antimicrobial hydrogels: promising materials for medical application. Int J Nanomedicine. 2018;13:2217–2263. | ||
Gao W, Chen Y, Zhang Y, Zhang Q, Zhang L. Nanoparticle-based local antimicrobial drug delivery. Adv Drug Deliv Rev. 2018;127:46–57. | ||
Dos Santos Ramos MA, Da Silva PB, Spósito L, et al. Nanotechnology-based drug delivery systems for control of microbial biofilms: a review. Int J Nanomedicine. 2018;13:1179–1213. | ||
Hibbitts A, O’Leary C. Emerging nanomedicine therapies to counter the rise of methicillin-resistant Staphylococcus aureus. Materials (Basel). 2018;11(2):E321. | ||
Khurana C, Chudasama B. Nanoantibiotics: strategic assets in the fight against drug-resistant superbugs. Int J Nanomedicine. 2018;13:3–6. | ||
Liu W, das J, Mepham AH, Nemr CR, Sargent EH, Kelley SO. A fully-integrated and automated testing device for PCR-free viral nucleic acid detection in whole blood. Lab Chip. 2018;18(13):1928–1935. | ||
Cheng D, Yu M, Fu F, et al. Dual recognition strategy for specific and sensitive detection of bacteria using aptamer-coated magnetic beads and antibiotic-capped gold nanoclusters. Anal Chem. 2016;88(1):820–825. | ||
Sung YJ, Suk HJ, Sung HY, Li T, Poo H, Kim MG. Novel antibody/gold nanoparticle/magnetic nanoparticle nanocomposites for immunomagnetic separation and rapid colorimetric detection of Staphylococcus aureus in milk. Biosens Bioelectron. 2013;43:432–439. | ||
Kumar N, Kulkarni K, Behera L, Verma V. Preparation and characterization of maghemite nanoparticles from mild steel for magnetically guided drug therapy. J Mater Sci Mater Med. 2017;28(8):116. | ||
Kearns H, Goodacre R, Jamieson LE, Graham D, Faulds K. SERS detection of multiple antimicrobial-resistant pathogens using nanosensors. Anal Chem. 2017;89(23):12666–12673. | ||
Cowger TA, Yang Y, Rink DE, et al. Protein-adsorbed magnetic-nanoparticle-mediated assay for rapid detection of bacterial antibiotic resistance. Bioconjug Chem. 2017;28(4):890–896. | ||
Wei X, Zhou W, Sanjay ST, et al. Multiplexed instrument-free bar-chart spinchip integrated with nanoparticle-mediated magnetic aptasensors for visual quantitative detection of multiple pathogens. Anal Chem. 2018;90(16):9888–9896. | ||
Gupta AK, Naregalkar RR, Vaidya VD, Gupta M. Recent advances on surface engineering of magnetic iron oxide nanoparticles and their biomedical applications. Nanomedicine (Lond). 2007;2(1):23–39. | ||
Corchero JL, Villaverde A. Biomedical applications of distally controlled magnetic nanoparticles. Trends Biotechnol. 2009;27(8):468–476. | ||
Chung HJ, Castro CM, Im H, Lee H, Weissleder R. A magneto-DNA nanoparticle system for rapid detection and phenotyping of bacteria. Nat Nanotechnol. 2013;8(5):369–375. | ||
Park KS, Kim H, Kim S, et al. Nanomagnetic system for rapid diagnosis of acute infection. ACS Nano. 2017;11(11):11425–11432. | ||
Diekema DJ, Beach ML, Pfaller MA, Jones RN; SENTRY Participants Group. Antimicrobial resistance in viridans group streptococci among patients with and without the diagnosis of cancer in the USA, Canada and Latin America. Clin Microbiol Infect. 2001;7(3):152–157. | ||
Bubeck Wardenburg J, Williams WA, Missiakas D. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc Natl Acad Sci U S A. 2006;103(37):13831–13836. | ||
Kaim AH, Wischer T, O’Reilly T, et al. MR imaging with ultrasmall superparamagnetic iron oxide particles in experimental soft-tissue infections in rats. Radiology. 2002;225(3):808–814. | ||
Liu J, Bai R, Li Y, et al. MRI detection of bacterial brain abscesses and monitoring of antibiotic treatment using bacCEST. Magn Reson Med. 2018;80(2):662–671. | ||
Hoerr V, Tuchscherr L, Hüve J, et al. Bacteria tracking by in vivo magnetic resonance imaging. BMC Biol. 2013;11(1):63. | ||
Moro L, Turemis M, Marini B, Ippodrino R, Giardi MT. Better together: strategies based on magnetic particles and quantum dots for improved biosensing. Biotechnol Adv. 2017;35(1):51–63. | ||
Veigas B, Fernandes AR, Baptista PV. AuNPs for identification of molecular signatures of resistance. Front Microbiol. 2014;5:455. | ||
Law JW, Ab Mutalib NS, Chan KG, Lee LH. Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front Microbiol. 2014;5:770. | ||
Kumar N, Hu Y, Singh S, Mizaikoff B. Emerging biosensor platforms for the assessment of water-borne pathogens. Analyst. 2018;143(2):359–373. | ||
Mocan T, Matea CT, Pop T, et al. Development of nanoparticle-based optical sensors for pathogenic bacterial detection. J Nanobiotechnology. 2017;15(1):25. | ||
Amendola V, Pilot R, Frasconi M, Maragò OM, Iatì MA. Surface plasmon resonance in gold nanoparticles: a review. J Phys Condens Matter. 2017;29(20):203002. | ||
Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277(5329):1078–1081. | ||
Storhoff JJ, Marla SS, Bao P, et al. Gold nanoparticle-based detection of genomic DNA targets on microarrays using a novel optical detection system. Biosens Bioelectron. 2004;19(8):875–883. | ||
Chan WS, Tang BS, Boost MV, Chow C, Leung PH. Detection of methicillin-resistant Staphylococcus aureus using a gold nanoparticle-based colourimetric polymerase chain reaction assay. Biosens Bioelectron. 2014;53:105–111. | ||
Wu S-Y, Hulme J, An SSA. Recent trends in the detection of pathogenic Escherichia coli O157:H7. BioChip J. 2015;9(3):173–181. | ||
Tsai TT, Huang CY, Chen CA, et al. Diagnosis of tuberculosis using colorimetric gold nanoparticles on a paper-based analytical device. ACS Sens. 2017;2(9):1345–1354. | ||
Cheon SA, Cho HH, Kim J, Lee J, Kim HJ, Park TJ. Recent tuberculosis diagnosis toward the end TB strategy. J Microbiol Methods. 2016;123:51–61. | ||
Tang L, Li J. Plasmon-based colorimetric nanosensors for ultrasensitive molecular diagnostics. ACS Sens. 2017;2(7):857–875. | ||
Foubert A, Beloglazova NV, Rajkovic A, et al. Bioconjugation of quantum dots: review and impact on future application. TrAC Trends Anal Chem. 2016;83:31–48. | ||
Goldman ER, Clapp AR, Anderson GP, et al. Multiplexed toxin analysis using four colors of quantum dot fluororeagents. Anal Chem. 2004;76(3):684–688. | ||
Banerjee R, Jaiswal A. Recent advances in nanoparticle-based lateral flow immunoassay as a point-of-care diagnostic tool for infectious agents and diseases. Analyst. 2018;143(9):1970–1996. | ||
Morales-Narváez E, Naghdi T, Zor E, Merkoçi A. Photoluminescent lateral-flow immunoassay revealed by graphene oxide: highly sensitive paper-based pathogen detection. Anal Chem. 2015;87(16):8573–8577. | ||
Jin B, Wang S, Lin M, et al. Upconversion nanoparticles based FRET aptasensor for rapid and ultrasenstive bacteria detection. Biosens Bioelectron. 2017;90:525–533. | ||
Date AA, Hanes J, Ensign LM. Nanoparticles for oral delivery: design, evaluation and state-of-the-art. J Control Release. 2016;240:504–526. | ||
Brooks BD, Brooks AE. Therapeutic strategies to combat antibiotic resistance. Adv Drug Deliv Rev. 2014;78:14–27. | ||
Laverman P, Dams ET, Storm G, et al. Microscopic localization of PEG-liposomes in a rat model of focal infection. J Control Release. 2001;75(3):347–355. | ||
Tran N, Hocquet M, Eon B, et al. Non-lamellar lyotropic liquid crystalline nanoparticles enhance the antibacterial effects of rifampicin against Staphylococcus aureus. J Colloid Interface Sci. 2018;519:107–118. | ||
Kaleko M, Bristol JA, Hubert S, et al. Development of SYN-004, an oral beta-lactamase treatment to protect the gut microbiome from antibiotic-mediated damage and prevent Clostridium difficile infection. Anaerobe. 2016;41:58–67. | ||
Dickson RP, Erb-Downward JR, Huffnagle GB. The role of the bacterial microbiome in lung disease. Expert Rev Respir Med. 2013;7(3):245–257. | ||
Spicer CD, Jumeaux C, Gupta B, Stevens MM. Peptide and protein nanoparticle conjugates: versatile platforms for biomedical applications. Chem Soc Rev. 2018;47(10):3574–3620. | ||
Chen H, Li M, Liu Z, et al. Design of antibacterial peptide-like conjugated molecule with broad spectrum antimicrobial ability. Sci China Chem. 2018;61(1):113–117. | ||
Zhao X, Pan F, Xu H, et al. Molecular self-assembly and applications of designer peptide amphiphiles. Chem Soc Rev. 2010;39(9):3480–3498. | ||
Mitra RN, Shome A, Paul P, Das PK. Antimicrobial activity, biocompatibility and hydrogelation ability of dipeptide-based amphiphiles. Org Biomol Chem. 2009;7(1):94–102. | ||
Radovic-Moreno AF, Lu TK, Puscasu VA, Yoon CJ, Langer R, Farokhzad OC. Surface charge-switching polymeric nanoparticles for bacterial cell wall-targeted delivery of antibiotics. ACS Nano. 2012;6(5):4279–4287. | ||
Cao B, Xiao F, Xing D, Hu X. Polyprodrug antimicrobials: remarkable membrane damage and concurrent drug release to combat antibiotic resistance of methicillin-resistant Staphylococcus aureus. Small. 2018;14(41):e1802008. | ||
Liu P, Xu G, Pranantyo D, Xu LQ, Neoh K-G, Kang E-T. pH-sensitive zwitterionic polymer as an antimicrobial agent with effective bacterial targeting. ACS Biomater Sci Eng. 2018;4(1):40–46. | ||
Mankoci S, Kaiser RL, Sahai N, Barton HA, Joy A. Bactericidal peptidomimetic polyurethanes with remarkable selectivity against Escherichia coli. ACS Biomater Sci Eng. 2017;3(10):2588–2597. | ||
Jadhav M, Kalhapure RS, Rambharose S, et al. Novel lipids with three C18-fatty acid chains and an amino acid head group for pH-responsive and sustained antibiotic delivery. Chem Phys Lipids. 2018;212:12–25. | ||
Chu L, Gao H, Cheng T, et al. A charge-adaptive nanosystem for prolonged and enhanced in vivo antibiotic delivery. Chem Commun (Camb). 2016;52(37):6265–6268. | ||
Gillies ER, Fréchet JM. pH-Responsive copolymer assemblies for controlled release of doxorubicin. Bioconjug Chem. 2005;16(2):361–368. | ||
Kalhapure RS, Sikwal DR, Rambharose S, et al. Enhancing targeted antibiotic therapy via pH responsive solid lipid nanoparticles from an acid cleavable lipid. Nanomedicine. 2017;13(6):2067–2077. | ||
Wang J, Byrne JD, Napier ME, DeSimone JM. More effective nanomedicines through particle design. Small. 2011;7(14):1919–1931. | ||
Hejazi R, Amiji M. Stomach-specific anti-H. pylori therapy; part III: effect of chitosan microspheres crosslinking on the gastric residence and local tetracycline concentrations in fasted gerbils. Int J Pharm. 2004;272(1–2):99–108. | ||
Jing ZW, Jia YY, Wan N, et al. Design and evaluation of novel pH-sensitive ureido-conjugated chitosan/TPP nanoparticles targeted to Helicobacter pylori. Biomaterials. 2016;84:276–285. | ||
Luo M, Jia YY, Jing ZW, et al. Construction and optimization of pH-sensitive nanoparticle delivery system containing PLGA and UCCs-2 for targeted treatment of Helicobacter pylori. Colloids Surf B Biointerfaces. 2018;164:11–19. | ||
Khutoryanskiy VV. Beyond PEGylation: alternative surface-modification of nanoparticles with mucus-inert biomaterials. Adv Drug Deliv Rev. 2018;124:140–149. | ||
Hayden SC, Zhao G, Saha K, et al. Aggregation and interaction of cationic nanoparticles on bacterial surfaces. J Am Chem Soc. 2012;134(16):6920–6923. | ||
Westmeier D, Posselt G, Hahlbrock A, et al. Nanoparticle binding attenuates the pathobiology of gastric cancer-associated Helicobacter pylori. Nanoscale. 2018;10(3):1453–1463. | ||
Yu H, Guo C, Feng B, et al. Triple-layered pH-responsive micelleplexes loaded with siRNA and cisplatin prodrug for NF-Kappa B targeted treatment of metastatic breast cancer. Theranostics. 2016;6(1):14–27. | ||
Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268(5219):1899–1902. | ||
Su Y, Zhao L, Meng F, Wang Q, Yao Y, Luo J. Silver nanoparticles decorated lipase-sensitive polyurethane micelles for on-demand release of silver nanoparticles. Colloids Surf B Biointerfaces. 2017;152:238–244. | ||
Chen YL, Zhu S, Zhang L, et al. Smart conjugated polymer nanocarrier for healthy weight loss by negative feedback regulation of lipase activity. Nanoscale. 2016;8(6):3368–3375. | ||
Xu L, He C, Hui L, et al. Bactericidal dendritic polycation cloaked with stealth material via lipase-sensitive intersegment acquires neutral surface charge without losing membrane-disruptive activity. ACS Appl Mater Interfaces. 2015;7(50):27602–27607. | ||
Yang S, Han X, Yang Y, et al. Bacteria-targeting nanoparticles with microenvironment-responsive antibiotic release to eliminate intracellular Staphylococcus aureus and associated infection. ACS Appl Mater Interfaces. 2018;10(17):14299–14311. | ||
Rezaee R, Talebreza A, Ziari K, Behnod V, Emampour BFS. Distribution of virulence factors and antimicrobial resistance properties of uropathogenic Escherichia coli isolated from diabetic and healthy males suffered from urinary tract infections. Biosci Biotechnol Res Asia. 2016;13(2):931–937. | ||
Lim YM, de Groof AJC, Bhattacharjee MK, Figurski DH, Schon EA. Bacterial conjugation in the cytoplasm of mouse cells. Infect Immun. 2008;76(11):5110–5119. | ||
Marshall TG, Lee RE, Marshall FE. Common angiotensin receptor blockers may directly modulate the immune system via VDR, PPAR and CCR2b. Theor Biol Med Model. 2006;3(1):1. | ||
Pires-Lapa M, Carvalho-Sousa C, Cecon E, Fernandes P, Markus R. β-adrenoceptors trigger melatonin synthesis in phagocytes. Int J Mol Sci. 2018;19(8):E2182. | ||
Proal AD, Albert PJ, Marshall TG, Blaney GP, Lindseth IA. Immunostimulation in the treatment for chronic fatigue syndrome/myalgic encephalomyelitis. Immunol Res. 2013;56(2–3):398–412. | ||
Aksungur P, Demirbilek M, Denkbaş EB, Vandervoort J, Ludwig A, Unlü N. Development and characterization of Cyclosporine A loaded nanoparticles for ocular drug delivery: cellular toxicity, uptake, and kinetic studies. J Control Release. 2011;151(3):286–294. | ||
Mohammadi G, Nokhodchi A, Barzegar-Jalali M, et al. Physicochemical and anti-bacterial performance characterization of clarithromycin nanoparticles as colloidal drug delivery system. Colloids Surf B Biointerfaces. 2011;88(1):39–44. | ||
Giannavola C, Bucolo C, Maltese A, et al. Influence of preparation conditions on acyclovir-loaded poly-d,l-lactic acid nanospheres and effect of PEG coating on ocular drug bioavailability. Pharm Res. 2003;20(4):584–590. | ||
Couvreur P, Fattal E, Alphandary H, Puisieux F, Andremont A. Intracellular targeting of antibiotics by means of biodegradable nanoparticles. J Control Release. 1992;19(1–3):259–267. | ||
Ranjan A, Pothayee N, Seleem M, et al. Drug delivery using novel nanoplexes against a Salmonella mouse infection model. J Nanopart Res. 2010;12(3):905–914. | ||
Sosnik A, Carcaboso AM, Glisoni RJ, Moretton MA, Chiappetta DA. New old challenges in tuberculosis: potentially effective nanotechnologies in drug delivery. Adv Drug Deliv Rev. 2010;62(4–5):547–559. | ||
Rawal T, Butani S. Combating tuberculosis infection: a forbidding challenge. Indian J Pharm Sci. 2016;78(1):8–16. | ||
Chew NY, Chan HK. Use of solid corrugated particles to enhance powder aerosol performance. Pharm Res. 2001;18(11):1570–1577. | ||
Lawlor C, Kelly C, O’Leary S, et al. Cellular targeting and trafficking of drug delivery systems for the prevention and treatment of MTb. Tuberculosis (Edinb). 2011;91(1):93–97. | ||
Pandey R, Sharma A, Zahoor A, Sharma S, Khuller GK, Prasad B. Poly (DL-lactide-co-glycolide) nanoparticle-based inhalable sustained drug delivery system for experimental tuberculosis. J Antimicrob Chemother. 2003;52(6):981–986. | ||
Fang G, Li W, Shen X, et al. Differential Pd-nanocrystal facets demonstrate distinct antibacterial activity against Gram-positive and Gram-negative bacteria. Nat Commun. 2018;9(1):129. | ||
Choi SK, Myc A, Silpe JE, et al. Dendrimer-based multivalent vancomycin nanoplatform for targeting the drug-resistant bacterial surface. ACS Nano. 2013;7(1):214–228. | ||
Yu M, Wang H, Fu F, et al. Dual-recognition Förster resonance energy transfer based platform for one-step sensitive detection of pathogenic bacteria using fluorescent vancomycin-gold nanoclusters and aptamer-gold nanoparticles. Anal Chem. 2017;89(7):4085–4090. | ||
Silvero CMJ, Rocca DM, de La Villarmois EA, et al. Selective photoinduced antibacterial activity of amoxicillin-coated gold nanoparticles: from one-step synthesis to in vivo cytocompatibility. ACS Omega. 2018;3(1):1220–1230. | ||
Zhu M, Liu W, Liu H, et al. Construction of Fe3O4/Vancomycin/PEG magnetic nanocarrier for highly efficient pathogen enrichment and gene sensing. ACS Appl Mater Interfaces. 2015;7(23):12873–12881. | ||
Wang C, Zhang K, Zhou Z. Vancomycin-modified Fe(3)O(4)@SiO(2)@Ag microflowers as effective antimicrobial agents. Int J Nanomedicine. 2017;12:3077–3094. | ||
Capeletti LB, Loiola LMD, Picco AS, da Silva Liberato M, Cardoso MB. 8 – Silica nanoparticle applications in the biomedical field. In: Ciofani G, editor. Smart Nanoparticles for Biomedicine. Elsevier; 2018:115–129. | ||
Kim Y, Park EJ, Na DH. Recent progress in dendrimer-based nanomedicine development. Arch Pharm Res. 2018;41(6):571–582. | ||
Wu S, Duan N, Qiu Y, Li J, Wang Z. Colorimetric aptasensor for the detection of Salmonella enterica serovar typhimurium using ZnFe2O4-reduced graphene oxide nanostructures as an effective peroxidase mimetics. Int J Food Microbiol. 2017;261:42–48. | ||
Angsantikul P, Thamphiwatana S, Zhang Q, et al. Coating nanoparticles with gastric epithelial cell membrane for targeted antibiotic delivery against Helicobacter pylori infection. Adv Ther (Weinh). 2018;1(2):1800016. | ||
Muppidi K, Wang J, Betageri G, Pumerantz AS. PEGylated liposome encapsulation increases the lung tissue concentration of vancomycin. Antimicrob Agents Chemother. 2011;55(10):4537–4542. | ||
Kalhapure RS, Sonawane SJ, Sikwal DR, et al. Solid lipid nanoparticles of clotrimazole silver complex: an efficient nano antibacterial against Staphylococcus aureus and MRSA. Colloids Surf B Biointerfaces. 2015;136:651–658. | ||
Pei Y, Mohamed MF, Seleem MN, Yeo Y. Particle engineering for intracellular delivery of vancomycin to methicillin-resistant Staphylococcus aureus (MRSA)-infected macrophages. J Control Release. 2017;267:133–143. | ||
Puisney C, Baeza-Squiban A, Boland S. Mechanisms of uptake and translocation of nanomaterials in the lung. Adv Exp Med Biol. 2018;1048:21–36. | ||
Berçot B, Poirel L, Dortet L, Nordmann P. In vitro evaluation of antibiotic synergy for NDM-1-producing Enterobacteriaceae. J Antimicrob Chemother. 2011;66(10):2295–2297. | ||
Johnson TJ, Siek KE, Johnson SJ, Nolan LK. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J Bacteriol. 2006;188(2):745–758. | ||
Sercombe L, Veerati T, Moheimani F, Wu SY, Sood AK, Hua S. Advances and challenges of liposome assisted drug delivery. Front Pharmacol. 2015;6(127):286. | ||
Deol P, Khuller GK, Joshi K. Therapeutic efficacies of isoniazid and rifampin encapsulated in lung-specific stealth liposomes against Mycobacterium tuberculosis infection induced in mice. Antimicrob Agents Chemother. 1997;41(6):1211–1214. | ||
Ruckmani K, Sankar V, Sivakumar M. Tissue distribution, pharmacokinetics and stability studies of zidovudine delivered by niosomes and proniosomes. J Biomed Nanotechnol. 2010;6(1):43–51. | ||
Huang CT, Stewart PS. Reduction of polysaccharide production in Pseudomonas aeruginosa biofilms by bismuth dimercaprol (BisBAL) treatment. J Antimicrob Chemother. 1999;44(5):601–605. | ||
Mahdiun F, Mansouri S, Khazaeli P, Mirzaei R. The effect of tobramycin incorporated with bismuth-ethanedithiol loaded on niosomes on the quorum sensing and biofilm formation of Pseudomonas aeruginosa. Microb Pathog. 2017;107:129–135. | ||
Chitambar CR. Gallium and its competing roles with iron in biological systems. Biochim Biophys Acta. 2016;1863(8):2044–2053. | ||
Hammer ND, Skaar EP. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu Rev Microbiol. 2011;65(1):129–147. | ||
Oh SH, Park HS, Kim HS, et al. Antimicrobial activities of LCB10-0200, a novel siderophore cephalosporin, against the clinical isolates of Pseudomonas aeruginosa and other pathogens. Int J Antimicrob Agents. 2017;50:700–706. | ||
Goderska K, Agudo Pena S, Alarcon T. Helicobacter pylori treatment: antibiotics or probiotics. Appl Microbiol Biotechnol. 2018;102(1):1–7. | ||
Trifan A, Girleanu I, Cojocariu C, et al. Pseudomembranous colitis associated with a triple therapy for Helicobacter pylori eradication. World J Gastroenterol. 2013;19(42):7476–7479. | ||
Miernyk KM, Bulkow LR, Gold BD, et al. Prevalence of Helicobacter pylori among Alaskans: factors associated with infection and comparison of urea breath test and anti-Helicobacter pylori IgG antibodies. Helicobacter. 2018;23(3):e12482. | ||
Wang YK, Kuo FC, Liu CJ, et al. Diagnosis of Helicobacter pylori infection: current options and developments. World J Gastroenterol. 2015;21(40):11221–11235. | ||
Ramteke S, Ganesh N, Bhattacharya S, Jain NK. Triple therapy-based targeted nanoparticles for the treatment of Helicobacter pylori. J Drug Target. 2008;16(9):694–705. | ||
Camargo MC, García A, Riquelme A, et al. The problem of Helicobacter pylori resistance to antibiotics: a systematic review in Latin America. Am J Gastroenterol. 2014;109(4):485–495. | ||
Li XX, Shi S, Rong L, Feng MQ, Zhong L. The impact of liposomal linolenic acid on gastrointestinal microbiota in mice. Int J Nanomedicine. 2018;13:1399–1409. | ||
Mullane K. Fidaxomicin in Clostridium difficile infection: latest evidence and clinical guidance. Ther Adv Chronic Dis. 2014;5(2):69–84. | ||
Xu K, Liang ZC, Ding X, et al. Nanomaterials in the prevention, diagnosis, and treatment of Mycobacterium tuberculosis infections. Adv Healthc Mater. 2018;7(1):1700509. | ||
Ai X, Hu M, Wang Z, et al. Recent advances of membrane-cloaked nanoplatforms for biomedical applications. Bioconjug Chem. 2018;29(4):838–851. | ||
Hu CM, Fang RH, Luk BT, Zhang L. Polymeric nanotherapeutics: clinical development and advances in stealth functionalization strategies. Nanoscale. 2014;6(1):65–75. | ||
Escajadillo T, Olson J, Luk BT, Zhang L, Nizet V. A red blood cell membrane-camouflaged nanoparticle counteracts streptolysin O-mediated virulence phenotypes of invasive Group A Streptococcus. Front Pharmacol. 2017;8:477. | ||
Charoenphol P, Oswalt K, Bishop CJ. Therapeutics incorporating blood constituents. Acta Biomater. 2018;73:64–80. | ||
Zhang Y, Zhang J, Chen W, et al. Erythrocyte membrane-coated nanogel for combinatorial antivirulence and responsive antimicrobial delivery against Staphylococcus aureus infection. J Control Release. 2017;263:185–191. | ||
Chen Y, Chen M, Zhang Y, et al. Broad-spectrum neutralization of pore-forming toxins with human erythrocyte membrane-coated nanosponges. Adv Healthc Mater. 2018;7(13):e1701366. | ||
Wei X, Gao J, Wang F, et al. In situ capture of bacterial toxins for antivirulence vaccination. Adv Mater. 2017;29(33):1701644. | ||
Fang RH, Luk BT, Hu CM, Zhang L. Engineered nanoparticles mimicking cell membranes for toxin neutralization. Adv Drug Deliv Rev. 2015;90:69–80. | ||
Jiang Y, Fang RH, Zhang L. Biomimetic nanosponges for treating antibody-mediated autoimmune diseases. Bioconjug Chem. 2018;29(4):870–877. | ||
Yuk SA, Sanchez-Rodriguez DA, Tsifansky MD, Yeo Y. Recent advances in nanomedicine for sepsis treatment. Ther Deliv. 2018;9(6):435–450. | ||
Das S, Angsantikul P, Le C, et al. Neutralization of cholera toxin with nanoparticle decoys for treatment of cholera. PLoS Negl Trop Dis. 2018;12(2):e0006266. | ||
Gao W, Fang RH, Thamphiwatana S, et al. Modulating antibacterial immunity via bacterial membrane-coated nanoparticles. Nano Lett. 2015;15(2):1403–1409. | ||
Shen H, Ackerman AL, Cody V, et al. Enhanced and prolonged cross-presentation following endosomal escape of exogenous antigens encapsulated in biodegradable nanoparticles. Immunology. 2006;117(1):78–88. | ||
Song C, Noh YW, Lim YT. Polymer nanoparticles for cross-presentation of exogenous antigens and enhanced cytotoxic T-lymphocyte immune response. Int J Nanomedicine. 2016;11:3753–3764. | ||
Bafica A, Scanga CA, Feng CG, Leifer C, Cheever A, Sher A. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J Exp Med. 2005;202(12):1715–1724. | ||
Weiss DS, Raupach B, Takeda K, Akira S, Zychlinsky A. Toll-like receptors are temporally involved in host defense. J Immunol. 2004;172(7):4463–4469. | ||
Lu D, Garcia-Contreras L, Xu D, et al. Poly (lactide-co-glycolide) microspheres in respirable sizes enhance an in vitro T cell response to recombinant Mycobacterium tuberculosis antigen 85B. Pharm Res. 2007;24(10):1834–1843. | ||
Cai H, Hu XD, Yu DH, Li SX, Tian X, Zhu YX. Combined DNA vaccine encapsulated in microspheres enhanced protection efficacy against Mycobacterium tuberculosis infection of mice. Vaccine. 2005;23(32):4167–4174. | ||
Priyadarsini S, Mukherjee S, Mishra M. Nanoparticles used in dentistry: a review. J Oral Biol Craniofac Res. 2018;8(1):58–67. | ||
Kadiyala U, Turali-Emre ES, Bahng JH, Kotov NA, VanEpps JS. Unexpected insights into antibacterial activity of zinc oxide nanoparticles against methicillin resistant Staphylococcus aureus (MRSA). Nanoscale. 2018;10(10):4927–4939. | ||
De Matteis V. Exposure to inorganic nanoparticles: routes of entry, immune response, biodistribution and in vitro/in vivo toxicity evaluation. Toxics. 2017;5(4):E29. | ||
Breznan D, das DD, O’Brien JS, et al. Differential cytotoxic and inflammatory potency of amorphous silicon dioxide nanoparticles of similar size in multiple cell lines. Nanotoxicology. 2017;11(2):223–235. | ||
Jain AK, Senapati VA, Singh D, Dubey K, Maurya R, Pandey AK. Impact of anatase titanium dioxide nanoparticles on mutagenic and genotoxic response in Chinese hamster lung fibroblast cells (V-79): the role of cellular uptake. Food Chem Toxicol. 2017;105:127–139. | ||
Vimbela GV, Ngo SM, Fraze C, Yang L, Stout DA. Antibacterial properties and toxicity from metallic nanomaterials. Int J Nanomedicine. 2017;12:3941–3965. | ||
Panáček A, Smékalová M, Kilianová M, et al. Strong and nonspecific synergistic antibacterial efficiency of antibiotics combined with silver nanoparticles at very low concentrations showing no cytotoxic effect. Molecules. 2015;21(1):E26. | ||
Thorsing M, Bentin T, Givskov M, Tolker-Nielsen T, Goltermann L. The bactericidal activity of β-lactam antibiotics is increased by metabolizable sugar species. Microbiology. 2015;161(10):1999–2007. | ||
Rahman T, Yarnall B, Doyle DA. Efflux drug transporters at the forefront of antimicrobial resistance. Eur Biophys J. 2017;46(7):647–653. | ||
Stokes JM, MacNair CR, Ilyas B, et al. Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat Microbiol. 2017;2(5):17028. | ||
Melander RJ, Melander C. The challenge of overcoming antibiotic resistance: an adjuvant approach? ACS Infect Dis. 2017;3(8):559–563. | ||
Wu SY, Park GY, Kim SH, Hulme J, An SSA. Diminazene aceturate: an antibacterial agent for Shiga-toxin-producing Escherichia coli O157:H7. Drug Des Devel Ther. 2016;10:3363–3378. | ||
Milton ME, Minrovic BM, Harris DL, et al. Re-sensitizing multidrug resistant bacteria to antibiotics by targeting bacterial response regulators: characterization and comparison of interactions between 2-aminoimidazoles and the response regulators BfmR from Acinetobacter baumannii and QseB from Francisella spp. Front Mol Biosci. 2018;5:15. | ||
Jamil B, Habib H, Abbasi SA, Ihsan A, Nasir H, Imran M. Development of cefotaxime impregnated chitosan as nano-antibiotics: de novo strategy to combat biofilm forming multi-drug resistant pathogens. Front Microbiol. 2016;7:330. | ||
Meng J, He G, Wang H, et al. Reversion of antibiotic resistance by inhibiting mecA in clinical methicillin-resistant Staphylococci by antisense phosphorothioate oligonucleotide. J Antibiot. 2015;68(3):158–164. | ||
Bikard D, Euler CW, Jiang W, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32(11):1146–1150. | ||
Luo ML, Leenay RT, Beisel CL. Current and future prospects for CRISPR-based tools in bacteria. Biotechnol Bioeng. 2016;113(5):930–943. | ||
Giau VV, Lee H, Shim KH, Bagyinszky E, An SSA. Genome-editing applications of CRISPR-Cas9 to promote in vitro studies of Alzheimer’s disease. Clin Interv Aging. 2018;13:221–233. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

