Back to Archived Journals » Advances in Genomics and Genetics » Volume 5
Peripheral blood mononuclear cell gene expression in healthy adults rapidly transported to high altitude
Authors Herman N, Grill D, Anderson P, Miller A, Johnson J, O'Malley K, Ceridon Richert M, Johnson B
Received 25 April 2014
Accepted for publication 17 June 2014
Published 16 December 2014 Volume 2015:5 Pages 1—9
DOI https://doi.org/10.2147/AGG.S66784
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Nicole M Herman,1 Diane E Grill,2 Paul J Anderson,1 Andrew D Miller,1 Jacob B Johnson,1 Kathy A O’Malley,1 Maile L Ceridon Richert,1 Bruce D Johnson1
1Department of Cardiovascular Diseases, 2Department of Biostatistics, Mayo Clinic Rochester, MN, USA
Abstract: Although mechanisms of high altitude illness have been studied extensively, the processes behind the development of these conditions are still unclear. Few genome-wide studies on rapid exposure to high altitude have been performed. Each year, scientists and support workers are transferred by plane from McMurdo Station in Antarctica (sea level) to the Amundsen-Scott South Pole Station at 2,835 meters. This uniform and rapid transfer to altitude provides a unique opportunity to study the effects of hypobaric hypoxia on gene expression that may help illustrate the body's adaptations to these conditions. We hypothesized that an extensive number of genes would change with rapid exposure to altitude and further expected that these genes would correspond to inflammatory pathways proposed as a mechanism in development of acute mountain sickness. Peripheral venous blood samples were drawn from 98 healthy subjects at sea level and again on day two at altitude. Microarray analysis was performed on these samples. In total, 1,118 probe sets with significant P-values and fold changes (90% upregulated) were identified and entered into MetaCore™ software. Several pathways, including oxidative phosphorylation, cytoskeleton remodeling, and platelet aggregation, were significantly represented by the data set and all were upregulated. Many genes changed expression, and the vast majority of these increased. Increased metabolism in peripheral blood mononuclear cells suggests increased inflammatory activity.
Keywords: peripheral blood mononuclear cells, microarray, gene expression, acute mountain sickness
Introduction
Exposure to high altitude results in a number of physiological changes that occur in an attempt to adapt or compensate for the reduced partial pressure of alveolar oxygen. This includes sympathetic activation, changes in water balance, and primary stimulation of peripheral chemoreceptors leading to increased minute ventilation.1
True acclimatization occurs over the course of many days to weeks, and includes a host of adaptive processes that permit the human body to function in a reduced-oxygen environment, including an increase in glycolytic metabolism2,3 and altered mitochondrial activity and protein synthesis.4 However, there may also be maladaptive responses that contribute to altitude-related illnesses. One example is an increased inflammatory response.5 The general consensus on minimizing the risk of developing altitude-related illness is to climb high but sleep low and to limit the daily increase in altitude to 300 m or a rest day for every 900 m of elevation gain.6,7 Typically, the latter is reserved for altitudes above 3,000 m.8
However, there are an increasing number of places that require more rapid transport without formal acclimatization paradigms. One example is the US Antarctic Program’s South Pole Station. This program requires a large number of personnel (250–300) to be flown in from sea level for the Antarctic summer, which lasts 100 days. Winter crews at the South Pole station are smaller, with 50–60 people. The South Pole is at the cusp of the altitude zone where symptoms begin to occur (2,835 m), and, due to lower than expected barometric pressure, the physiological altitude is higher than the physical altitude.
Personnel who work at the South Pole Station make the ascent during a short 3–4-hour flight from the sea level Antarctic base at McMurdo Station. We have previously reported that there is a high incidence (52%) of acute mountain sickness (AMS) in subjects making this transition,9 that physiological measures of body water regulation and inflammation are key in development of AMS,10 and that oral contraceptive use increases symptoms of AMS.11 Thus, the ascent profile and infrastructure of the US Antarctic Program offers a unique opportunity to capture large numbers of healthy subjects exposed rapidly to altitude in a uniform fashion to better understand the adaptive and maladaptive physiological mechanisms involved.
The mechanisms of AMS remain unclear and, while the responses of a given individual to a given ascent may be variable, there appear to be individuals more susceptible to developing altitude symptoms, implying some degree of genetic predisposition. However, there are few studies assessing predisposition to AMS in families.1,12 A number of genes and specific polymorphisms are believed to be associated with high altitude exposure, such as HIF-1, the ACE I/D polymorphism, the Arg16/Gly polymorphism of the β2 adrenergic receptor, NOS3, and HSP70.1 It has been proposed that AMS is the result of changes in several mechanistic pathways, including central and peripheral sympathetic nervous system activation, the renin–angiotensin pathway and water balance, activation of free radicals, development of oxidative stress, and inflammatory processes,1 as well as vasogenic changes in cerebral fluid balance and metabolism.13,14
However, no single physiological pathway has been conclusively associated with susceptibility to AMS, and many of the polymorphisms thought to be associated with development of the disease have not consistently been shown to correlate with development of AMS.1 The vast majority of studies exploring these genes in relation to hypoxia are studies of polymorphic alleles in genes of interest that have already been identified and the correlation of these alleles with the incidence of AMS. Studies using muscle biopsies for analysis report significant decreases in mitochondrial activity. Glycolytic metabolism, lipid peroxidation, and expression of mitochondrial uncoupling proteins increase during subacute hypoxia, and the proportions of muscle fiber types are altered, presumably to protect cells from the reactive oxygen species produced by mitochondria in hypoxic conditions. Over time, there is a decrease in the number of mitochondria, along with expression of uncoupling proteins and certain components of the electron transport chain. These adaptations take place over weeks and months of exposure to hypobaric hypoxia.15,16 Investigation of a small number of genes and specific polymorphisms involves significant limitations. With tens of thousands of genes in the human genome and far more potential polymorphisms, investigating 20 genes and one or two polymorphisms per gene cannot account for all possible significant variations. Techniques like microarray analysis and high-throughput sequencing offer opportunities to investigate genome-wide changes and identify new genes and pathways of interest.
Microarrays are small chips with embedded oligonucleotide sequences (probes) that are complementary to conserved regions of specific genes. Complementary DNA produced from isolated RNA is applied to the chip, and the probe fluoresces in proportion to the quantity of DNA bound.17 Comparing the relative fluorescence of these probes between samples provides a way of assessing changes in gene expression. Some studies have performed microarray analysis using peripheral blood mononuclear cells (PBMCs) to study gene expression in different disease states involving inflammatory processes such as heart failure,2 sepsis,18 inflammatory arthritis,19 type 1 diabetes,20 cancer,21 and systemic infections.22,23 Because of the proposed inflammatory component in altitude illness, using PBMCs may provide an important insight into the physiological responses of the body to acute hypobaric hypoxia.
In this study, we took advantage of the controlled transport of large numbers of healthy adults to high altitude, and identified genes that were differentially expressed. In addition, we explored potential pathways involving these genes in an attempt to gain an understanding of the cellular adaptations that occur in response to altitude and specific genes that may change with acute altitude exposure. We hypothesized that an extensive array of genes would show increased expression with rapid transport to altitude, and we anticipated that these genes would correspond to predefined pathways related primarily to inflammatory processes.
Materials and methods
Overview
As previously reported,9 the study was conducted with funding from the National Science Foundation, and data were collected in the summer seasons of 2006–2007 and 2007–2008 (October to November). The study protocol was approved by the institutional review board at Mayo Clinic in Rochester, MN, USA. All subjects gave written consent after reviewing the study protocol and receiving brief education on high altitude illness, and all aspects of the study conformed to the Declaration of Helsinki.9 Personnel arriving at McMurdo Station at sea level in Antarctica answered a questionnaire, underwent simple physiological tests, and had blood samples taken to determine baseline measures of biochemical intermediates. All subjects were offered prophylactic acetazolamide (Diamox®) through the US Antarctic Program if they wished to take it. A subset of 98 of these subjects recruited sequentially from the larger study group had blood drawn also for gene expression analysis, and these subjects constitute the cohort for this study. The majority of subjects stayed at the McMurdo Station for a period of 2 weeks prior to transport to the South Pole. After transport to the South Pole, questionnaires regarding health status and AMS were administered at five time points at the same time of day, and another blood sample was taken after two nights at altitude.
Gene expression data collection
Blood samples were collected at McMurdo Station and on the third day at the South Pole Station in PAXgene Blood RNA tubes (PreAnalytiX, Hombrechtikon, Switzerland). The tubes were then frozen at −40°C until their return to the USA. Following transport to the USA, the samples were thawed and the cells pelleted by centrifugation. RNA was extracted using Rneasy MiniKits (Qiagen, Valencia, CA, USA) and stored at −80°C. Microarray analysis was performed using the Affymetrix GeneChip Human Genome U133 Plus 2 array. This technology analyzes the expression level of more than 47,000 transcripts and variants with more than 54,000 probe sets. The sequences used in the probe sets were derived from sequences from GenBank, dbEST, and RefSeq. Arrays were run in the Mayo Clinic Advanced Genomics Technology Center to quantify the data used in the analysis. The data discussed in this publication have been deposited in the Gene Expression Omnibus at the National Center for Biotechnology Information and are accessible through Gene Expression Omnibus series accession number GSE46480.
Statistical methods
The Affymetrix array contains 54,675 probe sets that typically contain eleven probes per probe set. Analyses were done using R statistical software (http://www.r-project.org/) and preprocessing was done using the RMA package from Bioconductor.24–28 Specifically, the data were normalized with quantile normalization and without background correction, and then summarized using Tukey’s median polish procedure.29 All analyses were done using the log2 of normalized fluorescence. The Wilcoxon signed-rank test was used to test for significant differences in gene expression between base camp and altitude. Average fold changes were calculated for each gene. The probe sets with either a fold change >log2(1.5) or a fold change <log2(1/1.5) and with P<0.05 were deemed to be statistically significant, giving 1,118 significant probe sets used in the pathway analysis. A Wilcoxon rank sum test was performed to assess the difference in expression between those who did and did not take acetazolamide during the study. A histogram was created to show the distribution of P-values for the probe sets (data not shown).
Pathway analysis
Thomson Reuters MetaCore™ GeneGO software (http://thomsonreuters.com/products_services/science/science_products/a-z/metacore/) was used for the enrichment and pathway analysis. In the enrichment analysis, gene IDs are matched to functional ontology sets in MetaCore™. GeneGO annotaters edit and maintain canonical pathway maps that represent about 650 metabolic and signaling pathways identified in human biology. Sets of gene IDs from experimental data sets are compared with the sets of IDs in canonical pathway maps, and the probability that the intersection of the experimental and established sets would be randomly assembled is estimated using a hypergeometric distribution. Smaller P-values indicate a higher relevance to the dataset, and pathways are ranked based on this P-value. In the pathway graphics, blue indicates downregulation of a given gene and red indicates upregulation.30
Results
Subject demographics
A total of 246 subjects participated in the ASAP (Antarctic Study of Altitude Physiology) study (mean age 37±11 years, 70% male, body mass index 26.1±4.2 kg/m2). All had been medically examined and determined to be healthy prior to arrival at McMurdo Station. Eighty-nine percent of subjects had prior exposure to high altitude and 61% of these subjects reported previous altitude illness.9 Of the 246 subjects, 98 (mean age 37.32±9.89 years, 70% male, body mass index 26.1±4.2, Table 1) had peripheral blood drawn for analysis of gene expression, and these subjects were included in this analysis. Of these 98 subjects, 33 elected to take prophylactic acetazolamide. Since 93% of the probe sets were not significantly different (P>0.05) between the group of subjects who had taken acetazolamide and the group that had not, the data are presented as a single group.
  | Table 1 Subject demographics |
Environmental and physiological data with rapid exposure to high altitude
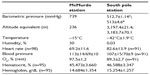
Information on environmental conditions and equivalent altitude is provided in Table 2. Upon transition to altitude, resting heart rate increased (from 69.2±11.6 bpm to 82.6±13.9 bpm, P<0.001) and oxygen saturation decreased (from 97.5%±1.2% to 89.3%±2.7%, P<0.001, Table 2). Hematocrit and hemoglobin both increased slightly, consistent with a small reduction in plasma volume due to dehydration.
Gene expression and pathway data
Of the 1,118 significant genes identified, 1,004 were upregulated and 114 were downregulated. The 20 genes with the most significant P-values (8.46E–18<P<1.12E–17) for the change in expression are shown in Table 3. Two of the probe sets were not associated with established human genes.
  | Table 3 Probe sets with most significant P-values in the data set |
The MetaCore™ enrichment analysis identified ten metabolic pathways that had the most significant representation of the selected probe sets (Table 4). The most significant pathway identified was oxidative phosphorylation (P=1.04E–20; Figure 1). Of the 105 genes identified in MetaCore™ as playing a role in this pathway, 26 were found in the experimental data set (Table 5). Two of the identified pathway components of MetaCore™ could not be directly related back to specific probe sets and therefore have no associated fold changes. All identified genes in this pathway are significantly upregulated.
  | Table 4 Pathways identified by MetaCore™ represented by the data set |
  | Table 5 Genes in the oxidative phosphorylation pathway present in the data set |
Discussion
Overview
PBMCs were isolated from blood samples drawn from 98 healthy workers traveling rapidly and in a uniform fashion from McMurdo Station at sea level in Antarctica to the South Pole Station located at 2,835 m. Messenger RNA was extracted for use in microarray gene expression analysis. A group of probe sets determined to be significant according to the P-value and fold change cutoffs described above was entered into the MetaCore software for pathway analysis. MetaCore identified ten pathways deemed to be most significantly represented by the restricted group of probes.
Primary findings
Overall, we observed a striking increase in a number of cellular processes associated with PBMC metabolism. The 20 probe sets with the most significant P-values from the Wilcoxon signed-rank test included genes associated with energy metabolism (UQCRQ, UQCR11, COX7B), translation and protein folding (RPL26, RPS24, PFDN5, CLEC2B), and cell growth and apoptosis (HUWE1, TPT1, BNIP3L). All of these genes showed statistically significant upregulation from baseline, suggesting an increase in PBMC metabolism. This would be consistent with proposed increases in PBMC activity and associated inflammation during exposure to high altitude. Increases in inflammatory cytokine secretion during hypoxia and other stresses have been well documented, along with corresponding activation of mononuclear cells.5,31 Eosinophils also secrete proinflammatory cytokines, prostaglandins, and leukotrienes, and increases in eosinophil activity also suggest an increase in inflammatory activity during hypoxia.32 The increase in activity in the identified pathways, particularly oxidative phosphorylation, cytoskeleton remodeling, regulation of initiation of translation, and signaling cascades, also indicates potential increases in PBMC activity. Genes shared by more than one pathway include PRKA, MLCP, ROCK, and NDUF. These genes are all significantly upregulated and play roles in energy and protein metabolism. Only three genes identified in the pathways, ie, AKAP13, NCOA1, and STAT3, were downregulated.
During acute hypoxia, the body compensates by attempting to increase oxygen delivery to the tissues. When tissues remain deficient in oxygen, cellular damage can occur when excessive amounts of free radicals are produced during metabolic processes such as oxidative phosphorylation, and the inflammatory response may be activated by the release of proinflammatory cytokines secreted in response to cell damage and death. The inflammatory response can then contribute to increased hypoxia in the tissues.33 Respiratory alkalosis from exposure to hypobaric hypoxia and cell death activate NFκB, which can trigger an inflammatory response. This inflammatory response and the associated increase in metabolic activity can in turn contribute to additional free radical production.34 Antioxidants such as vitamin E may help mitigate this effect, and prophylactic administration of antioxidants has been shown to reduce symptoms of AMS in a few studies.35 Upregulation of PBMCs has been observed in conditions associated with chronic inflammation and has been associated with other proinflammatory markers in the blood.36,37 Decreases in expression of genes involved in oxidative phosphorylation, ubiquinone metabolism, cell adhesion, and mitogen-activated protein kinase signaling have been correlated with decreases in reactive oxygen species markers occurring after dietary intervention.37 We observed a significant increase in gene expression associated with these pathways, and this is consistent with an increase in systemic inflammation.
Studies of gene expression changes related to hypoxia have often used tissue biopsies to investigate changes in tissues such as skeletal muscle.4,16 However, the extreme environment and field conditions present in this study made biopsies difficult to store, ship, and process. Because of the minimally invasive sampling technique, PBMCs were used in the microarray analysis. To our knowledge, PBMCs have not been used to evaluate gene expression in acute exposure to hypobaric hypoxia, although they have been used for analysis in other conditions. PBMCs are a heterogeneous group of cells, and different patterns of gene expression in each cell type can decrease the significance of the results.38 However, the changes in our data are very significant, with P-values in the range of 10−18 throughout the probe sets determined to be significant and used for analysis.
We believe that no previous studies have looked at gene expression in PBMCs with rapid exposure to altitude in a uniform protocol. Our results are novel and make an important contribution to the existing knowledge regarding gene expression changes with hypobaric hypoxia. The majority of studies involving gene expression analysis and altitude have investigated expression in other tissues, such as muscle, and assessed changes that occur with subchronic and chronic hypoxia exposure in conditions such as heart failure. These studies report downregulation of many genes and processes, particularly those associated with metabolism, including oxidative phosphorylation and protein synthesis.4,16 The differences in tissues and exposure times make these disparate results potentially compatible with ours. Additional studies will need to be performed to determine the relationship between hypoxia responses in tissues including PBMCs and muscle. Collecting samples from different tissues over several time points and a longer exposure period may help clarify the physiological responses to altitude in different areas of the body.
Implications
Inflammation is believed to play a role in the development of altitude illness. This is supported by the efficacy of anti-inflammatory drugs such as ibuprofen, dexamethasone, and budesonide in preventing symptoms of AMS and other high altitude illnesses in subjects traveling to high altitude.39–43 Nonsteroidal anti-inflammatory drugs like ibuprofen decrease inflammation by inhibiting production of prostaglandins and thromboxanes during an inflammatory response.43 Dexamethasone, a potent glucocorticoid, binds to the glucocorticoid receptor, downregulating the immune system by influencing the transcription of inflammatory proteins, most notably interleukin-1β, and modulating sympathetic activity to relieve symptoms of AMS.40–42,44 The glucocorticoid signaling pathway was upregulated in our data set, suggesting altered steroid metabolism at altitude. The exact mechanism by which inflammation contributes to the development of symptoms of AMS, such as headache and nausea, is not clear, although increased vascular permeability and vasodilation in the brain have been proposed as potential causes of these symptoms.11–14,41,43 Hypoxia and inflammation are mutually reinforcing in the body. Cellular damage due to increased metabolic stress triggers an inflammatory response, and the increased use of oxygen by immune cells that are recruited to the tissue contributes further to hypoxia. These processes result in changes to the epithelium that may eventually lead to development of AMS.33 As discussed in the Introduction, other conditions, such as chronic heart failure, chronic obstructive pulmonary disease, sleep apnea, and systemic infections, also result in increased inflammation. Investigating the role of inflammation in altitude illness may translate into a greater understanding of the role of inflammation in these conditions.
Acetazolamide, a standard preventive treatment for AMS, alters fluid balance by influencing the bicarbonate balance in the blood. It has been proposed that the diuretic effect of acetazolamide may help to prevent the symptoms that arise due to increased vascular permeability and accumulation of fluid in tissues.45 Several subjects in this study elected to take prophylactic acetazolamide, and we intend to investigate how acetazolamide may have influenced changes in gene expression in these subjects.
The upregulation observed in genes associated with metabolism in PBMCs in our data set supports the important role of increased inflammatory activity in development of AMS. We are further investigating how gene expression relates to the presence of AMS in subjects to understand important pathways better, not just in regards to genes with the greatest changes but also in regards to the genes most associated with development of AMS in a subject.
Limitations
We investigated how gene expression was altered in PBMCs in healthy adults rapidly transported to high altitude in a uniform fashion. A truly uniform protocol for the movement from sea level to high altitude ensured that all subjects experienced the same ascent profile. The cohort consisted of healthy adults with no potentially confounding medical conditions such as cardiovascular disease. Subjects were recruited sequentially and without regard to prior exposure to hypoxia or clinical presentation. The number of subjects (n=98) was large and allowed for a statistically significant sample size. Despite the potential for subject self-selection, all subjects served as their own controls in the analysis.
There are several important limitations to note. As discussed above, PBMCs represent a heterogeneous tissue, and gene expression may differ between specific cell types. The results seen here are specific to this tissue and cannot be generalized to other tissues types, such as muscle. Assessment of gene expression in only one cell type limits the conclusions that can be drawn about the greater picture of physiological changes occurring in response to hypoxia, including cytokine signaling and the inflammatory response. Second, the relationship between gene expression and production of the finished protein varies greatly for different genes in different cell types. Many genes, including HIF-1, are regulated at the post-transcriptional level; microarray analysis is unable to capture changes in post-transcriptional modification and may not provide a complete picture of the resulting phenotype. Third, some subjects elected to take prophylactic acetazolamide, and this is a potentially significant confounding factor. Withholding access to acetazolamide was considered unethical for US Antarctic Program personnel, and thus the gene expression data presented include all subjects, regardless of use of acetazolamide. The nature of the effect of acetazolamide on this data set is currently being investigated and will be explored in future papers. An estimated 7% of probe sets were significantly different between the two groups, but the physiological relevance of these differences is currently not known. Fourth, although transport to the South Pole was uniform, other aspects of lifestyle such as diet were not controlled. There were differences in the temperature and humidity between McMurdo Station at sea level and the South Pole Station, but subjects spent the majority of their time in climate-controlled environments or with appropriately issued extreme weather gear, so it would not be expected that core temperatures would have been influenced.
Conclusion
We found that rapid exposure to high altitude provokes gene upregulation in PBMC metabolism. This upregulation may play a role in the inflammation associated with altitude illness. In a study of 98 subjects who were transported uniformly to high altitude, PBMCs were isolated and gene expression was analyzed using microarray technology. Gene pathways including oxidative phosphorylation and cytoskeletal remodeling were identified as being significantly represented by probe sets, with significant fold changes and P-values. This upregulation is consistent with the increases in inflammatory activity believed to play a role in the development of AMS. Further analysis will be performed relating significant probe sets to the presence of AMS in each subject and identifying important sets of genes using principal component analysis.
Acknowledgments
We would like to thank the hard working employees of the US Antarctic Program who each year make their way to the South Pole to support the many scientific research agendas in Antarctica. This includes general science support, South Pole station management and cargo crew, Crary Laboratory, the medical team, and transportation/shipping at both McMurdo and the South Pole. We would also like to thank Jay O’Brien for his help and support in this project in Antarctica.
Disclosure
The authors report no conflicts of interest in this work.
References
MacInnis MJ, Wang P, Koehle MS, Rupert JL. The genetics of altitude tolerance: the evidence for inherited susceptibility to acute mountain sickness. Int J Occup Environ Med. 2011;53(2):159−168. | |
Cappuzzello C, Napolitano M, Arcelli D, et al. Gene expression profiles in peripheral blood mononuclear cells of chronic heart failure patients. Physiol Genomics. 2009;38(3):233−240. | |
Kakinuma Y, Miyauchi T, Yuki K, Murakoshi N, Goto K, Yamaguchi I. Novel molecular mechanism of increased myocardial endothelin-1 expression in the failing heart involving the transcriptional factor hypoxia-inducible factor-1alpha induced for impaired myocardial energy metabolism. Circulation. 2001;103(19):2387−2394. | |
Vigano A, Ripamonti M, De Palma S, et al. Proteins modulation in human skeletal muscle in the early phase of adaptation to hypobaric hypoxia. Proteomics. 2008;8(22):4668−4679. | |
Hartmann A, Pfuhler S, Dennog C, Germadnik D, Pilger A, Speit G. Exercise-induced DNA effects in human leukocytes are not accompanied by increased formation of 8-hydroxy-2′-deoxyguanosine or induction of micronuclei. Free Radic Biol Med. 1998;24(2):245–251. | |
Hsu TY, Weng YM, Chiu YH, et al. Rate of ascent and acute mountain sickness at high altitude. Clin J Sport Med. April 18, 2014. [Epub ahead of print.] | |
McDevitt M, McIntosh SE, Rodway G, Peelay J, Adams DL, Kayser B. Risk determinants of acute mountain sickness in trekkers in the nepali himalaya: a 24-year follow-up. Wilderness Environ Med. 2014;25(2):152−159. | |
Schneider M, Bernasch D, Weymann J, Holle R, Bartsch P. Acute mountain sickness: influence of susceptibility, preexposure, and ascent rate. Med Sci Sports Exerc. 2002;34(12):1886−1891. | |
Anderson PJ, Miller AD, O’Malley KA, et al. Incidence and symptoms of high altitude illness in South Pole workers: Antarctic Study of Altitude Physiology (ASAP). Clin Med Insights Circ Respir Pulm Med. 2011;5:27−35. | |
Harrison MF, Anderson P, Miller A, et al. Physiological variables associated with the development of acute mountain sickness at the South Pole. BMJ Open. 2013;3(7). | |
Harrison MF, Anderson P, Miller A, et al. Oral contraceptive use and acute mountain sickness in South Pole workers. Aviat Space Environ Med. 2013;84(11):1166−1171. | |
Rupert JL, Koehle MS. Evidence for a genetic basis for altitude-related illness. High Alt Med Biol. 2006;7(2):150−167. | |
Bailey DM, Kleger GR, Holzgraefe M, Ballmer PE, Bartsch P. Pathophysiological significance of peroxidative stress, neuronal damage, and membrane permeability in acute mountain sickness. J Appl Physiol. 2004;96(4):1459−1463. | |
Krasney JA. A neurogenic basis for acute altitude illness. Med Sci Sports Exerc. 1994;26(2):195−208. | |
Raguso CA, Guinot SL, Janssens JP, Kayser B, Pichard C. Chronic hypoxia: common traits between chronic obstructive pulmonary disease and altitude. Curr Opin Clin Nutr Metab Care. 2004;7(4):411−417. | |
Levett DZ, Radford EJ, Menassa DA, et al. Acclimatization of skeletal muscle mitochondria to high-altitude hypoxia during an ascent of Everest. FASEB J. 2012;26(4):1431−1441. | |
Christie JD. Microarrays. Crit Care Med. 2005;33(Suppl 12):S449−S452. | |
Tang BM, McLean AS, Dawes IW, Huang SJ, Lin RC. Gene-expression profiling of peripheral blood mononuclear cells in sepsis. Crit Care Med. 2009;37(3):882−888. | |
Myles A, Tuteja A, Aggarwal A. Synovial fluid mononuclear cell gene expression profiling suggests dysregulation of innate immune genes in enthesitis-related arthritis patients. (Rheumatology, Oxford). 2012;51(10):1785−1789. | |
Wang Z, Zheng C, Tan YY, et al. Gene expression changes in patients with fulminant type 1 diabetes. Chin Med J (Engl). 2011;124(22):3613−3617. | |
Kossenkov AV, Vachani A, Chang C, et al. Resection of non-small cell lung cancers reverses tumor-induced gene expression changes in the peripheral immune system. Clin Cancer Res. 2011;17(18):5867−5877. | |
Kang SS, Kim HJ, Jang MS, et al. Gene expression profile of human peripheral blood mononuclear cells induced by Staphylococcus aureus lipoteichoic acid. Int Immunopharmacol. 2012;13(4):454−460. | |
Wahl-Jensen V, Kurz S, Feldmann F, et al. Ebola virion attachment and entry into human macrophages profoundly effects early cellular gene expression. PLoS Negl Trop Dis. 2011;5(10):e1359. | |
Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185−193. | |
Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307−315. | |
Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. | |
Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. | |
Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249−264. | |
Tukey JW. Exploratory Data Analysis. Reading, MA, USA: Addison-Wesley; 1977. | |
Ekins S, Nikolsky Y, Bugrim A, Kirillov E, Nikolskaya T. Pathway mapping tools for analysis of high content data. Methods Mol Biol. 2007;356:319−350. | |
Klokker M, Kharazmi A, Galbo H, Bygbjerg I, Pedersen BK. Influence of in vivo hypobaric hypoxia on function of lymphocytes, neutrocytes, natural killer cells, and cytokines. J Appl Physiol. 1993;74(3):1100−1106. | |
Nissim Ben Efraim AH, Eliashar R, Levi-Schaffer F. Hypoxia modulates human eosinophil function. Clin Mol Allergy. 2010;8:10. | |
Eltzschig HK, Carmeliet P. Hypoxia and inflammation. N Engl J Med. 2011;364(7):656−665. | |
Behn C, Araneda OF, Llanos AJ, Celedon G, Gonzalez G. Hypoxia-related lipid peroxidation: evidences, implications and approaches. Respir Physiol Neurobiol. 2007;158(2−3):143−150. | |
Bailey DM, Davies B. Acute mountain sickness; prophylactic benefits of antioxidant vitamin supplementation at high altitude. High Alt Med Biol. 2001;2(1):21−29. | |
Bouwens M, van de Rest O, Dellschaft N, et al. Fish-oil supplementation induces antiinflammatory gene expression profiles in human blood mononuclear cells. Am J Clin Nutr. 2009;90(2):415−424. | |
van Dijk SJ, Feskens EJ, Bos MB, et al. Consumption of a high monounsaturated fat diet reduces oxidative phosphorylation gene expression in peripheral blood mononuclear cells of abdominally overweight men and women. J Nutr. 2012;142(7):1219−1225. | |
Jacobsen M, Repsilber D, Gutschmidt A, et al. Deconfounding microarray analysis – independent measurements of cell type proportions used in a regression model to resolve tissue heterogeneity bias. Methods Inf Med. 2006;45(5):557−563. | |
Gertsch JH, Corbett B, Holck PS, et al. Altitude Sickness in Climbers and Efficacy of NSAIDs Trial (ASCENT): randomized, controlled trial of ibuprofen versus placebo for prevention of altitude illness. Wilderness Environ Med. 2012;23(4):307−315. | |
Johnson TS, Rock PB, Fulco CS, Trad LA, Spark RF, Maher JT. Prevention of acute mountain sickness by dexamethasone. N Engl J Med. 1984;310(11):683−686. | |
Maggiorini M, Brunner-La Rocca HP, Peth S, et al. Both tadalafil and dexamethasone may reduce the incidence of high-altitude pulmonary edema: a randomized trial. Ann Intern Med. 2006;145(7):497−506. | |
Zheng CR, Chen GZ, Yu J, et al. Inhaled budesonide and oral dexamethasone prevent acute mountain sickness: a double-blind randomized controlled trial. Am J Med. April 28, 2014. [Epub ahead of print.] | |
Lipman GS, Kanaan NC, Holck PS, Constance BB, Gertsch JH. Ibuprofen prevents altitude illness: a randomized controlled trial for prevention of altitude illness with nonsteroidal anti-inflammatories. Ann Emerg Med. 2012;59(6):484−490. | |
Zhang W, Smith C, Howlett C, Stanimirovic D. Inflammatory activation of human brain endothelial cells by hypoxic astrocytes in vitro is mediated by IL-1beta. J Cereb Blood Flow Metab. 2000;20(6):967−978. | |
Leaf DE, Goldfarb DS. Mechanisms of action of acetazolamide in the prophylaxis and treatment of acute mountain sickness. J Appl Physiol. 2007;102(4):1313−1322. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.