Back to Journals » Drug Design, Development and Therapy » Volume 9
Paeoniflorin inhibits human glioma cells via STAT3 degradation by the ubiquitin–proteasome pathway
Authors Nie X, Ou-yang J, Xing Y, Li D, Dong X, Liu R, Xu R
Received 6 August 2015
Accepted for publication 9 September 2015
Published 13 October 2015 Volume 2015:9 Pages 5611—5622
DOI https://doi.org/10.2147/DDDT.S93912
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Wei Duan
Xiao-hu Nie,1,* Jia Ou-yang,2,* Ying Xing,3 Dan-yan Li,4 Xing-yu Dong,1 Ru-en Liu,5 Ru-xiang Xu6
1Affiliated Bayi Brain Hospital, Southern Medical University, Beijing, People’s Republic of China; 2Nanchang University Medical College, Jiangxi, People’s Republic of China; 3Department of Gastroenterology, The 98th Hospital of Nanjing Military Command, Huzhou, Zhejiang, People’s Republic of China; 4Spleen & Stomach Institution, Guangzhou University of Traditional Chinese Medicine, Guangdong, People’s Republic of China; 5Department of Neurosurgery, China–Japan Friendship Hospital, Beijing, People’s Republic of China; 6Bayi Brain Hospital, The Military General Hospital of Beijing PLA, Beijing, People’s Republic of China
*These authors contributed equally to this work
Abstract: We investigated the underlying mechanism for the potent proapoptotic effect of paeoniflorin (PF) on human glioma cells in vitro, focusing on signal transducer and activator of transcription 3 (STAT3) signaling. Significant time- and dose-dependent apoptosis and inhibition of proliferation were observed in PF-treated U87 and U251 glioma cells. Expression of STAT3, its active form phosphorylated STAT3 (p-STAT3), and several downstream molecules, including HIAP, Bcl-2, cyclin D1, and Survivin, were significantly downregulated upon PF treatment. Overexpression of STAT3 induced resistance to PF, suggesting that STAT3 was a critical target of PF. Interestingly, rapid downregulation of STAT3 was consistent with its accelerated degradation, but not with its dephosphorylation or transcriptional modulation. Using specific inhibitors, we demonstrated that the prodegradation effect of PF on STAT3 was mainly through the ubiquitin–proteasome pathway rather than via lysosomal degradation. These findings indicated that PF-induced growth suppression and apoptosis in human glioma cells through the proteasome-dependent degradation of STAT3.
Keywords: paeoniflorin, glioma, apoptosis, proliferation, signal transducer and activator of transcription 3 (STAT3), ubiquitin–proteasome pathway (UPP)
Introduction
Gliomas are the most common primary tumors of the human central nervous system.1 Despite tremendous efforts to improve the prognosis of glioma patients over the past four decades, very little progress has been made, especially in patients with high-grade gliomas (grades III and IV).2 As in other cancers, glioma progression is accompanied by abnormal molecular changes.3,4 Several molecules are vital in the pro- or antisurvival pathways.5,6 Therefore, the identification and/or development of chemotherapy agents that selectively target molecular events linked to cancer progression may be an effective approach for the treatment of glioma.7
Signal transducer and activator of transcription 3 (STAT3) is a transcription factor that integrates signals from extracellular stimuli and regulates genes involved in many important cellular processes, such as cell cycle progression, apoptosis, and angiogenesis.8,9 The role of STAT3 as a crucial oncoprotein in tumorigenesis has been extensively studied.10–12 After activation, phosphorylated-STAT3 could induce expression of genes that participate in oncogenesis, such as apoptosis inhibitors (HIAP-1, Bcl-2, and Survivin), and cell-cycle regulators (cyclin D1).13,14 Its expression and activation have also been correlated with reduced survival and poor prognosis in patients with glioma.15,16 Therefore, STAT3 is considered a promising target for glioma therapy, and its inhibition using multiple approaches has been shown to induce growth arrest and apoptosis in glioma cells both in vitro and in vivo.17–19
Interest in the identification of novel anticancer agents derived from natural sources has been growing exponentially. These agents provide an alternative approach to improve the existing standard of care for cancer patients.20,21 Paeoniflorin (PF), an active component of Paeonia lactiflora Pall. (also called P. alba), has been widely used in traditional Chinese medicine. Previous studies have demonstrated that PF has many biological activities, such as immunoregulation,22,23 neuroprotection,24,25 and hepatoprotection.26 Recent research has indicated that PF might be a potent antitumor agent. It induced cell cycle arrest of HT29 colorectal cancer cells through the activation of p53/14-3-3 zeta.27 It also suppressed NF-κB activation and enhanced 5-fluorouracil-induced apoptosis in human gastric carcinoma cells.28 In this study, we demonstrated that PF inhibited proliferation and induced apoptosis of glioma cells. Additionally, we explored its antitumor activity via ubiquitylation and downregulation of STAT3. These attributes make PF a promising candidate compound for the treatment and prevention of glioma.
Materials and methods
Chemicals, reagents, antibodies, and cell culture
All human cell experiments were conducted according to the protocols approved by the Institutional Ethics Committee of Southern Medical University and the Military General Hospital of Beijing PLA. The study was approved by the Institutional Ethics Committee of Southern Medical University and Military General Hospital of Beijing PLA. DMSO (#D4540), Annexin V-FITC/PI apoptosis detection kit (#APOAF), proteasome inhibitor MG132 (#M7449), chloroquine (CQ) (#C6628), and thiazolyl blue tetrazolium bromide (MTT) (#M2128) were purchased from Sigma-Aldrich (St Louis, MO, USA). Protein A/G plus-agarose was obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA, #sc-2003). PF was purchased from Tianjin Shilan Technology (Tianjin, People’s Republic of China, #PCM-PL-002). PF diluted in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen, Valencia, CA, USA, #11965) was prepared as a stock solution of 400 mM and stored at −20°C until use. Antibodies specific for p-STAT3 (Tyr705) (#9145), STAT3 (#9139), cyclin D1 (#2978), Survivin (#2808), ubiquitin (#3936), and β-actin (#3700) were obtained from Cell Signaling Technology (Danvers, MA, USA). Human U87 and U251 cell lines were obtained from the Chinese Academy of Medical Sciences (Beijing, People’s Republic of China). Cells were maintained in DMEM supplemented with 10% fetal bovine serum (Invitrogen, #12664-025) in a humidified incubator with 5% CO2 at 37°C. The cells were passaged twice weekly and used for experiments when in the exponential growth phase.
Cell viability assay
Cells (4×103 cells per well) were seeded into 96-well plates in triplicate and incubated overnight before treatment with PF (10–20 mM) for 12–72 hours. After incubation, MTT dye was added, and the plates were incubated for 4 hours. Absorbance was measured using an ELISA reader (Multiskan EX; Labsystems; Dynex Technologies, Denkendorf, Germany) at a test wavelength of 490 nm and a reference wavelength of 690 nm.
Annexin V-FITC/PI staining
Approximately 1.5–2×105 cells per well were plated in six-well plates, treated with 10–20 mM PF for 24 hours or 20 mM for 6, 12, and 24 hours, then collected and stained with Annexin V-FITC/PI according to the manufacturer’s instructions (Sigma-Aldrich). Briefly, the cells were washed twice with cold phosphate-buffered saline (PBS), resuspended in binding buffer at 5×105 cells/500 μL, stained with 5 μL Annexin V and 10 μL Propidium iodide (PI), and incubated for 15 minutes at room temperature in the dark prior to flow cytometric analysis.
Western blot
Treated cells were washed in ice-cold PBS and extracted with ProteoJET™ Mammalian Cell Lysis Reagent (Fermentas, Burlington, ON, Canada) supplemented with protease and phosphatase inhibitors (Fermentas) according to the manufacturer’s instructions. Protein (20–40 μg) was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis using 8%–10% gels and then transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). Blots were blocked for 1 hour at room temperature with 5% bovine serum albumin (Sigma-Aldrich) in Tris-buffered saline/0.1% Tween-20. The blots were then incubated with specific primary antibodies at 4°C overnight. The blots were then incubated with horseradish peroxidase-linked secondary antibodies for 1 hour at room temperature. After additional washes, signals were detected using SuperSignal ECL (Pierce, Rockford, IL, USA).
Overexpression of wild-type STAT3
Semiconfluent cultures of U87 cells in 60 mm2 dishes or 96-well plates were transfected with indicated empty or STAT3 expression vector (pCMV6-STAT3) (Bioworld, Beijing, People’s Republic of China) using Lipofectamine™ 2000 reagent following the manufacturer’s protocol. Approximately 24 hours after transfection, cells were incubated with PF or vehicle for 24 hours. Overexpression of STAT3 in transfected cells was confirmed by immunoblotting.
RNA preparation and real-time PCR
Total RNA was extracted from U87 and U251 cells after treatment with PF for 24 hours using an E.Z.N.A. Total RNA Kit (Omega Bio-Tek, Norcross, GA, USA). RNA was then reverse-transcribed using a Prime-Script II 1st Strand cDNA Synthesis Kit (Takara, Shiga, Japan). For validation, real-time polymerase chain reaction (PCR) was performed using a SYBR Premix Ex Taq Kit (Takara) and an ABI Vii7 detection system (Applied Bio-systems, Foster City, CA, USA). The reaction conditions were: 95°C for 30 seconds, 95°C for 5 seconds, and 60°C for 34 seconds (40 cycles). The nucleotide sequences of the primers used for amplification were: STAT3 (forward, 5′-GCTTCCTGCAAGAGTCGAAT-3′; reverse, 5′-ATTGGCTTCTCAAGATACCTG-3′) and β-actin (forward, 5′-AGCGCGGCTACAGCTTCA-3′; reverse, 5′-GGCCATCTCTTGCTCGAAGT-3′).
Immunoprecipitation of STAT3 for ubiquitination assay
To assess the ubiquitin regulatory ability of PF in glioma, cells were treated with PF or vehicle for 24 hours, extracted with lysis buffer, and then centrifuged at 13,000× g for 20 minutes at 4°C. The cleared supernatants were removed, mixed with STAT3 antibody, and incubated for 1 hour at 4°C. Next, 20 μL of resuspended volume of Protein A/G PLUS-Agarose was added to the mixture, followed by incubation at 4°C on a rotating device overnight. The beads were collected by centrifugation at 10,000× g for 2 minutes and washed three times in cold PBS containing protease inhibitors and phosphatase inhibitors. The immunoprecipitation complex was subjected to SDS–PAGE, followed by immunoblotting with antiubiquitin antibody to detect polyubiquitinated STAT3 protein. Immune complexes were visualized using an enhanced chemiluminescence kit.
Statistical analysis
SPSS 13.0 software (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. Data were presented as the means ± SD. Statistical analysis of in vitro drug assays was performed by using the one-way ANOVA test and post hoc Bonferroni-corrected t-test. P<0.05 was considered statistically significant.
Results
Paeoniflorin inhibits human glioma cell proliferation
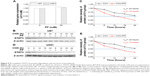
Cells were treated with different concentrations of PF for the indicated time periods, and MTT assay was used to measure cell viability. As shown in Figure 1A and B, cell viability was significantly decreased in a dose- and time-dependent manner in PF-treated cells (P<0.05). To assess the effect of PF on apoptosis, PF-treated cells stained with Annexin V-FITC/PI were analyzed by flow cytometry. Data shown in Figure 1C–F indicate that PF treatment increased apoptosis of glioma cells.
STAT3 mediates the role of paeoniflorin in glioma cell proliferation and apoptosis
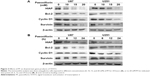
The ability of PF to modulate STAT3 in glioma cell lines was investigated. PF was found to downregulate both total STAT3 and pSTAT3 protein levels in a time- and dose-dependent manner (Figure 2A–H, P<0.05). Several signaling molecules downstream of STAT3, including HIAP, Bcl-2, cyclin D1, and Survivin, were also decreased by PF treatment in a time- and dose-dependent manner (Figure 3A and B). These findings confirm that PF modulates STAT3 signaling in U87 and U251 cells.
If STAT3 is a key target of PF, then forced overexpression of STAT3 should attenuate its antitumor effects. To test this hypothesis, we conducted a STAT3-overexpression experiment using cDNA of wild-type STAT3. The wild-type STAT3-overexpressing U87 cells showed increased expression of total and phosphorylated STAT3 (Figure 4A–D). STAT3 overexpression in U87 cells significantly attenuated the PF-mediated decrease in p-STAT3 (Figure 4C and D), indicating that the decrease of p-STAT3 might be due to STAT3 modulation. As expected, STAT3 overexpression attenuated the antiproliferative and proapoptotic effects of PF, in comparison with their respective control groups (Figure 4E–G). These findings suggest that PF might function, at least in part, via downregulation of STAT3 in glioma cells.
Paeoniflorin promotes ubiquitin-mediated STAT3 degradation
To investigate the mechanism by which PF decreases STAT3 expression, we first evaluated mRNA expression using real-time PCR after incubation with 20 mM PF or vehicle for 24 hours in both cell lines. There was no significant change in mRNA levels following PF treatment compared to the control group (Figure 5A). Next, using cycloheximide (CHX), an inhibitor of de novo protein synthesis, we showed that PF treatment influences STAT3 protein stability. As shown in Figure 5B–E, the half-life of STAT3 protein in cells treated with PF and CHX was much shorter (~12 hours in U87 and ~13 hours in U251, respectively) than in cells treated with CHX alone (~24 hours for both). Thus, the degradation of STAT3 was significantly accelerated when treated with PF and CHX. These results indicate that PF influences STAT3 protein stability without affecting its transcription in glioma cells.
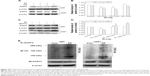
Both proteasomes and lysosomes are important mediators of degradation of cellular proteins. To determine which process was involved in PF-induced turnover of STAT3, cells were pretreated with the proteasomal inhibitor, MG132, or the lysosomal inhibitor, chloroquine (CQ), for 4 hours, followed by PF treatment or vehicle for an additional 20 hours. As shown in Figure 6A–D, lysosomal inhibition did not rescue PF-induced STAT3 downregulation. In contrast, MG132 pretreatment completely blocked PF-mediated degradation of STAT3 in both cell lines. We next performed an in vitro ubiquitination activity assay to examine the role of ubiquitination in PF-mediated proteasomal degradation of STAT3 in both glioma cell lines. Interestingly, degraded STAT3 could be detected using an antiubiquitin antibody, indicating that STAT3 was ubiquitinated during PF-induced degradation. Specifically, we demonstrated that the intensity of the smeared bands of STAT3 in PF-treated cells was stronger than that in control cells (Figure 6E). These results indicate that the ubiquitin–proteasome pathway (UPP) plays an important role in PF-mediated STAT3 degradation in glioma cells.
Discussion
In this study, we have shown that PF inhibits proliferation and induces apoptosis of human glioma cells. Importantly, we have provided evidence that PF regulates STAT3 turnover via the ubiquitin–proteasome system. Therefore, PF is the first natural product targeting STAT3 for ubiquitin-mediated proteasomal degradation.
STAT3 protein is present in the cytoplasm in an inactive form until it is phosphorylated by receptor-associated kinases. Activated JAK kinases phosphorylate STAT3 at the tyrosine residue in its SH2 domain. This phosphorylation event releases p-STAT3 from the receptor, allowing it to then homo- or heterodimerize with other STAT proteins. Dimerized STAT3 translocates to the nucleus and binds to promoters containing its cognate DNA-binding sequence. The targeting and disruption of oncogenic STAT3 signaling may be achieved via multiple approaches. Newly discovered chemotherapeutic reagents targeting STAT3 mainly act by inhibiting its tyrosine 705 phosphorylation.29–31 In this study, we found that PF strongly inhibits the STAT3 pathway. Specifically, PF decreases levels of total and phosphorylated STAT3 as well as several of its downstream targets. Unlike other chemotherapeutic agents that target the STAT3 pathway, PF did not directly inhibit phosphorylation at tyrosine 705. As shown in Figure 2, the expression levels of p-STAT3 protein correlated with those of STAT3. When STAT3 was overexpressed in PF-treated U87 cells, it rescued p-STAT3 protein level as well as cellular proliferation and antiapoptosis (Figure 4). These data clearly demonstrate that PF inhibits cell proliferation and induces apoptosis by suppressing STAT3 signaling, which was not mediated by dephosphorylation of STAT3, but by the levels of STAT3 itself.
In all tissues, proteins are mainly degraded via the UPP or by lysosomes. Lysosomes mainly degrade extracellular and some cell-surface proteins, while proteasomes degrade intracellular proteins that are aberrantly folded or normally short lived.32 These proteins are recognized by a specific E3 ubiquitin ligase and conjugated to polyubiquitin, which is then degraded by the proteasome in an ATP-dependent manner. These changes in the fates of individual proteins can, in turn, induce global changes in cell signaling, thus regulating cell proliferation and apoptosis. The clinical success of bortezomib, which targets the UPP-mediated degradation of proteins in cancer cells, illustrated the importance of ubiquitin-mediated signaling in cancer and stimulated further research in this field.33,34 Natural products can also trigger the ubiquitin-mediated turnover of some proteins, leading to antiproliferative and proapoptotic effects in cancer cells.35–39 A previous study showed that fucoidan, a polysaccharide extracted from brown seaweed, enhanced the UPP-mediated degradation of transforming growth factor receptor (TGFR) and reversed TGFR-induced epithelial-to-mesenchymal transition in breast cancer cell lines.40 In this study, STAT3 mRNA expression was not altered by PF treatment (Figure 5A). In addition, CHX and PF increased the rate of STAT3 degradation (Figure 5B–E). Therefore, the reduction of STAT3 by PF was not caused by synthesis inhibition, but by its accelerated degradation. MG132, a proteasome inhibitor, reversed STAT3 degradation (Figure 6C and D), whereas CQ, a known lysosome inhibitor, had no effect on STAT3 regulation (Figure 6A and B). We also detected increased ubiquitin–STAT3 complexes following PF treatment in both cell lines (Figure 6E).
Recently, Wang et al27 observed that PF inhibited the growth of HT29 colon carcinoma xenografts in nude mice. A dose of 1 g/kg PF/d caused no loss in bodyweight or diet consumption by oral gavage for 6 weeks compared with the control group. However, its inhibitory effects have not been evaluated yet. On the other hand, PF is an orally administered bioactive reagent that may act in the central nervous system, its ability to penetrate the blood–brain barrier should also be evaluated in future studies. Although we have demonstrated that PF promotes the UPP-mediated degradation of STAT3, the exact molecular mechanism is still unclear. As mentioned earlier, E3 ubiquitin ligase is a key player during UPPs, and many small molecules including some natural compounds specifically target E3 ubiquitin ligase to initiate the ubiquitin-mediated proteasomal degradation.36,41,42 As a result, we attempted to determine if any molecules (especially E3 ubiquitin ligase) were involved in PF-induced degradation of STAT3, but research in this field is limited.43,44 TMF/ARA160 and the E3 ligase TRAF6, which were reported to be involved in the UPP-mediated degradation of STAT3,43,44 were also investigated in this study (data not shown). Unfortunately, no significant differences were observed in their expression after PF treatment in two glioma cell lines.
Conclusion
In conclusion, we present the anticancer properties of PF in glioma cell lines. PF may achieve its antiproliferative and proapoptotic effects by reducing protein expression of STAT3 and its downstream targets. Furthermore, PF caused STAT3 degradation via the UPP. This is the first evidence that STAT3 is targeted for UPP-mediated degradation by a natural product. These data not only suggest that PF is a potential therapeutic candidate for glioma, but also demonstrate that the inhibition of STAT3 by its UPP-mediated degradation is an efficient strategy for cancer prevention and the discovery of new chemotherapeutic agents.
Acknowledgments
This work was supported by funding from the “Military Twelfth Five-Year Key Sci-Tech Research Projects” (Grant numbers BWS11J002 and BWS12J010).
Disclosure
The authors report no conflicts of interest in this work.
References
Ahmed R, Oborski MJ, Hwang M, Lieberman FS, Mountz JM. Malignant gliomas: current perspectives in diagnosis, treatment, and early response assessment using advanced quantitative imaging methods. Cancer Manag Res. 2014;6:149–170. | ||
Corwin D, Holdsworth C, Rockne RC, et al. Toward patient-specific, biologically optimized radiation therapy plans for the treatment of glioblastoma. PloS One. 2013;8(11):e79115. | ||
Fan W, Chen X, Li C, Chen L, Liu P, Chen Z. The analysis of deregulated expression and methylation of the PER2 genes in gliomas. J Cancer Res Ther. 2014;10(3):636–640. | ||
Kanamori M, Higa T, Sonoda Y, et al. Activation of the NRF2 pathway and its impact on the prognosis of anaplastic glioma patients. Neuro Oncol. 2015;17(4):555–565. | ||
Hu B, Emdad L, Bacolod MD, et al. Astrocyte elevated gene-1 interacts with Akt isoform 2 to control glioma growth, survival, and pathogenesis. Cancer Res. 2014;74(24):7321–7332. | ||
Cherry AE, Stella N. G protein-coupled receptors as oncogenic signals in glioma: emerging therapeutic avenues. Neuroscience. 2014;278C:222–236. | ||
Wurth R, Pattarozzi A, Gatti M, et al. Metformin selectively affects human glioblastoma tumor-initiating cell viability: a role for metformin-induced inhibition of Akt. Cell Cycle. 2013;12(1):145–156. | ||
Yue P, Turkson J. Targeting STAT3 in cancer: how successful are we? Expert Opin Investig Drugs. 2009;18(1):45–56. | ||
Siveen KS, Sikka S, Surana R, et al. Targeting the STAT3 signaling pathway in cancer: role of synthetic and natural inhibitors. Biochim Biophys Acta. 2014;1845(2):136–154. | ||
Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98(3):295–303. | ||
Timofeeva OA, Tarasova NI, Zhang X, et al. STAT3 suppresses transcription of proapoptotic genes in cancer cells with the involvement of its N-terminal domain. Proc Natl Acad Sci U S A. 2013;110(4):1267–1272. | ||
Luwor RB, Stylli SS, Kaye AH. The role of Stat3 in glioblastoma multiforme. J Clin Neurosci. 2013;20(7):907–911. | ||
Grivennikov SI, Karin M. Inflammation and oncogenesis: a vicious connection. Curr Opin Genet Dev. 2010;20(1):65–71. | ||
Kamran MZ, Patil P, Gude RP. Role of STAT3 in cancer metastasis and translational advances. Bio Med Res Int. 2013;2013:421821. | ||
Alvarez JV, Mukherjee N, Chakravarti A, et al. A STAT3 gene expression signature in gliomas is associated with a poor prognosis. Transl Oncogenomics. 2007;2:99–105. | ||
Caldera V, Mellai M, Annovazzi L, Valente G, Tessitore L, Schiffer D. Stat3 expression and its correlation with proliferation and apoptosis/autophagy in gliomas. J Oncol. 2008;2008:219241. | ||
Zhang H, Nie W, Zhang X, et al. NEDD4-1 regulates migration and invasion of glioma cells through CNrasGEF ubiquitination in vitro. PloS One. 2013;8(12):e82789. | ||
Ball S, Li C, Li PK, Lin J. The small molecule, LLL12, inhibits STAT3 phosphorylation and induces apoptosis in medulloblastoma and glioblastoma cells. PloS One. 2011;6(4):e18820. | ||
Rahaman SO, Harbor PC, Chernova O, Barnett GH, Vogelbaum MA, Haque SJ. Inhibition of constitutively active Stat3 suppresses proliferation and induces apoptosis in glioblastoma multiforme cells. Oncogene. 2002;21(55):8404–8413. | ||
Yu T, Zhou Z, Mu Y, de Lima Lopes G, Luo KQ. A novel anti-cancer agent, acetyltanshinone IIA, inhibits oestrogen receptor positive breast cancer cell growth by down-regulating the oestrogen receptor. Cancer Lett. 2014;346(1):94–103. | ||
Wang X, Duan X, Yang G, et al. Honokiol crosses BBB and BCSFB, and inhibits brain tumor growth in rat 9L intracerebral gliosarcoma model and human U251 xenograft glioma model. PloS One. 2011;6(4):e18490. | ||
Nizamutdinova IT, Jin YC, Kim JS, et al. Paeonol and paeoniflorin, the main active principles of Paeonia albiflora, protect the heart from myocardial ischemia/reperfusion injury in rats. Planta Med. 2008;74(1):14–18. | ||
Zheng YQ, Wei W, Zhu L, Liu JX. Effects and mechanisms of Paeoniflorin, a bioactive glucoside from paeony root, on adjuvant arthritis in rats. Inflamm Res. 2007;56(5):182–188. | ||
Mao QQ, Zhong XM, Feng CR, Pan AJ, Li ZY, Huang Z. Protective effects of paeoniflorin against glutamate-induced neurotoxicity in PC12 cells via antioxidant mechanisms and Ca(2+) antagonism. Cell Mol Neurobiol. 2010;30(7):1059–1066. | ||
Wang K, Zhu L, Zhu X, et al. Protective effect of paeoniflorin on Abeta25-35-induced SH-SY5Y cell injury by preventing mitochondrial dysfunction. Cell Mol Neurobiol. 2014;34(2):227–234. | ||
Liu DF, Wei W, Song LH. Protective effect of paeoniflorin on immunological liver injury induced by bacillus Calmette-Guerin plus lipopolysaccharide: modulation of tumour necrosis factor-alpha and interleukin-6 MRNA. Clin Exp Pharmacol Physiol. 2006;33(4):332–339. | ||
Wang H, Zhou H, Wang CX, et al. Paeoniflorin inhibits growth of human colorectal carcinoma HT 29 cells in vitro and in vivo. Food Chem Toxicol. 2012;50(5):1560–1567. | ||
Wu H, Li W, Wang T, Shu Y, Liu P. Paeoniflorin suppress NF-kappaB activation through modulation of I kappaB alpha and enhances 5-fluorouracil-induced apoptosis in human gastric carcinoma cells. Biomed Pharmacother. 2008;62(9):659–666. | ||
Gupta SC, Phromnoi K, Aggarwal BB. Morin inhibits STAT3 tyrosine 705 phosphorylation in tumor cells through activation of protein tyrosine phosphatase SHP1. Biochem Pharmacol. 2013;85(7):898–912. | ||
Kim SM, Lee JH, Sethi G, et al. Bergamottin, a natural furanocoumarin obtained from grapefruit juice induces chemosensitization and apoptosis through the inhibition of STAT3 signaling pathway in tumor cells. Cancer Lett. 2014;354(1):153–163. | ||
Rajendran P, Li F, Shanmugam MK, et al. Honokiol inhibits signal transducer and activator of transcription-3 signaling, proliferation, and survival of hepatocellular carcinoma cells via the protein tyrosine phosphatase SHP-1. J Cell Physiol. 2012;227(5):2184–2195. | ||
Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17(7):1807–1819. | ||
Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458(7237):438–444. | ||
Berkers CR, Ovaa H. Drug discovery and assay development in the ubiquitin-proteasome system. Biochem Soc Trans. 2010;38 (Pt 1):14–20. | ||
Shen M, Chan TH, Dou QP. Targeting tumor ubiquitin-proteasome pathway with polyphenols for chemosensitization. Anticancer Agents Med Chem. 2012;12(8):891–901. | ||
Shen SC, Lee WR, Yang LY, Tsai HH, Yang LL, Chen YC. Quercetin enhancement of arsenic-induced apoptosis via stimulating ROS-dependent p53 protein ubiquitination in human HaCaT keratinocytes. Exp Dermatol. 2012;21(5):370–375. | ||
Hsu HY, Lin TY, Wu YC, et al. Fucoidan inhibition of lung cancer in vivo and in vitro: role of the Smurf2-dependent ubiquitin proteasome pathway in TGFbeta receptor degradation. Oncotarget. 2014;5(17):7870–7885. | ||
Liu YB, Gao X, Deeb D, et al. Ubiquitin-proteasomal degradation of antiapoptotic survivin facilitates induction of apoptosis in prostate cancer cells by pristimerin. Int J Oncol. 2014;45(4):1735–1741. | ||
Samuel W, Kutty RK, Duncan T, et al. Fenretinide induces ubiquitin-dependent proteasomal degradation of stearoyl-CoA desaturase in human retinal pigment epithelial cells. J Cell Physiol. 2014;229(8):1028–1038. | ||
Hsu HY, Lin TY, Hwang PA, et al. Fucoidan induces changes in the epithelial to mesenchymal transition and decreases metastasis by enhancing ubiquitin-dependent TGFbeta receptor degradation in breast cancer. Carcinogenesis. 2013;34(4):874–884. | ||
Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat Rev Drug Dis. 2006;5(7):596–613. | ||
Jeong JH, An JY, Kwon YT, Li LY, Lee YJ. Quercetin-induced ubiquitination and down-regulation of Her-2/neu. J Cell Biochem. 2008;105(2):585–595. | ||
Perry E, Tsruya R, Levitsky P, et al. TMF/ARA160 is a BC-box-containing protein that mediates the degradation of Stat3. Oncogene. 2004;23(55):8908–8919. | ||
Wei J, Yuan Y, Jin C, et al. The ubiquitin ligase TRAF6 negatively regulates the JAK-STAT signaling pathway by binding to STAT3 and mediating its ubiquitination. PloS One. 2012;7(11):e49567. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.